Figures & data
Figure 1. Multialignment of the ι-CA amino acid sequences from different species (bacteria, cyanobacteria, diatoms, and algae). In red, the putative residues of the catalytic triad (T106, Y124, S199); light blue colour, the H197 as the putative proton shuttle residue. In green are the other residues of the catalytic pocket (see Hirakawa et al. [Citation64]). The putative motif (H)HHSS is the typical consensus sequence characterising the ι-CAs. The asterisk (*) indicates identity at all aligned positions. The multialignment was performed with MUSCLE, version 3.1. See for the identification of the amino acid sequences used in the multialignment. The residue number system used refers to the AspCA enzyme.
![Figure 1. Multialignment of the ι-CA amino acid sequences from different species (bacteria, cyanobacteria, diatoms, and algae). In red, the putative residues of the catalytic triad (T106, Y124, S199); light blue colour, the H197 as the putative proton shuttle residue. In green are the other residues of the catalytic pocket (see Hirakawa et al. [Citation64]). The putative motif (H)HHSS is the typical consensus sequence characterising the ι-CAs. The asterisk (*) indicates identity at all aligned positions. The multialignment was performed with MUSCLE, version 3.1. See Table 1 for the identification of the amino acid sequences used in the multialignment. The residue number system used refers to the AspCA enzyme.](/cms/asset/aa94b6d7-6e91-457b-b23f-fbc33a7a9635/ienz_a_1972995_f0001_c.jpg)
Figure 2. Phylogenetic analysis of ι-CAs from various organisms. The dendrogram was constructed using the ι-CA amino acid sequences reported in .
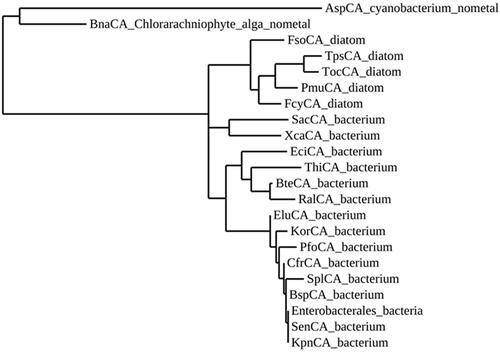
Table 1. Organisms, acronyms, and accession numbers of the amino acid sequences used in the phylogenetic analysis of ι-CAs.
Figure 3. Ribbon view of (A) hNFT2 (PDB 1GY5), (B) CAMKII in complex with acetate (PDB 2W2C), (C) AspCA (ι-CA from Anabaena cyanobacterium) in complex with HCO3- (PDB 7C5V), (D) BnaCA (ι-CA from microalga Bigelowiella natans) in complex with I- (PDB 7C5Y), (E) XcaCA (ι-CA from X. campestris) in complex with putative HCO3- (PDB 3H51), (F) homology model of BteCA (ι-CA from B. territorii) using XcaCA as template (51% identity).
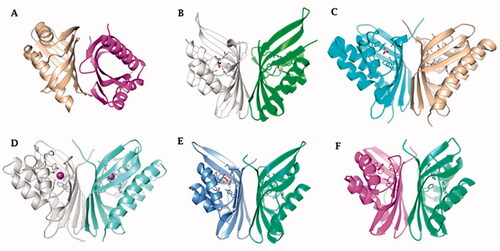
Figure 4. (A) Ribbon upper view and (B) binding site view of AspCA (ι-CA from Anabaena cyanobacterium) in complex with HCO3- (PDB 7C5V), (C) Ribbon upper view and (D) binding site view of XcaCA (ι-CA from X. campestris) in complex with putative HCO3- (PDB 3H51). Residue numbers according to AspCA (PDB 7C5V).
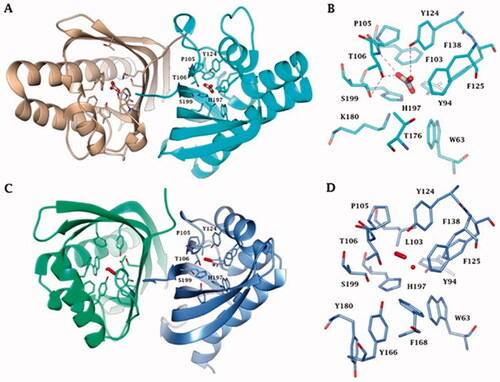
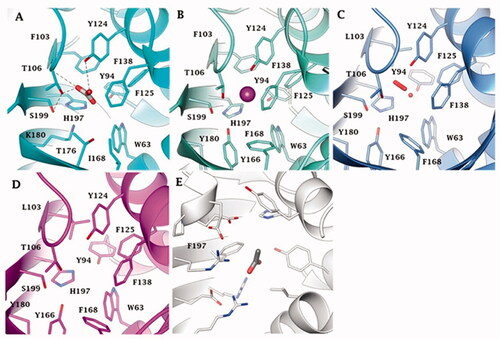
Figure 5. Binding site view of (A) AspCA (ι-CA from Anabaena cyanobacterium) in complex with HCO3- (PDB 7C5V), (B) BnaCA (ι-CA from microalga Bigelowiella natans) in complex with I- (PDB 7C5Y), (C) XcaCA (ι-CA from X. campestris) in complex with putative HCO3- (PDB 3H51), (D) homology model of BteCA (ι-CA from B. territorii) using XcaCA as template (51% identity), (E) CAMKII in complex with acetate (PDB 2W2C). Residue numbers according to AspCA (PDB 7C5V).