Figures & data
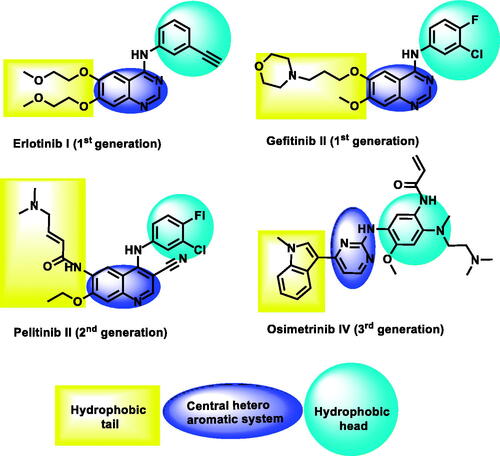
Figure 1. The essential pharmacophoric features of erlotinib as an EGFR inhibitor occupying three pockets in the ATP binding site based on ReferenceCitation22.
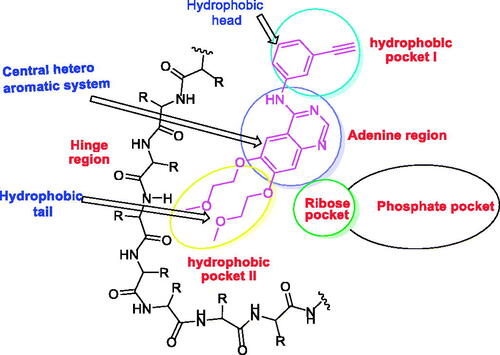
Table 1. Percentage of growth inhibition activity of compounds 7a, 8a–d, 9a, 10a–e against A549, PC-3, HCT-116 and MCF-7 at a concentration of 100 μM.
Table 2. IC50 values of compounds 8a, 8b, 8d, 9a and 12b against A-549, PC-3, HCT-116 and MCF-7.
Table 3. In vitro enzymatic inhibitory activities against EGFRL858R and EGFR790M.
Table 4. Effect of compound 8a on active caspase-3 in PC-3 cells after 24 h treatment.
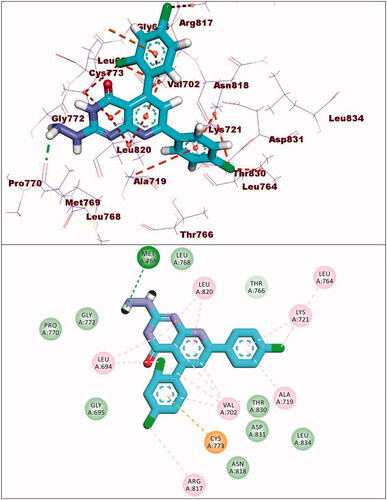
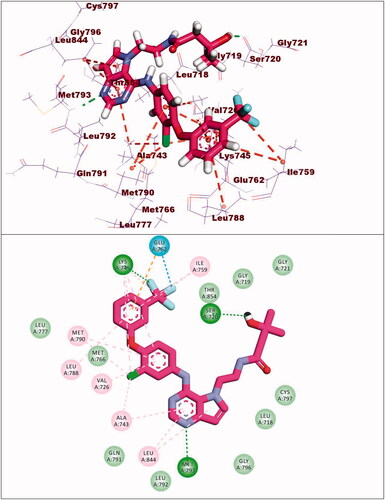
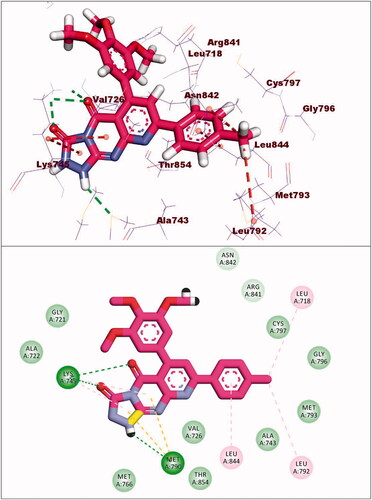
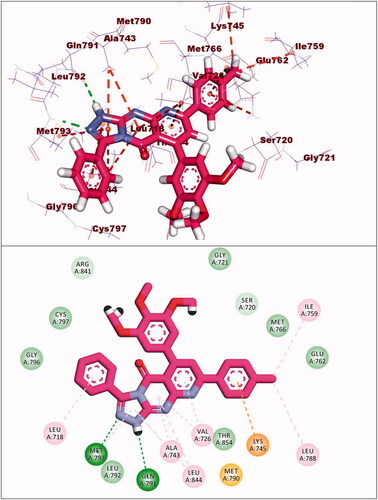
Table 5. The docking binding free energies of the synthesised compounds against EGFRWT and EGFRT790M.
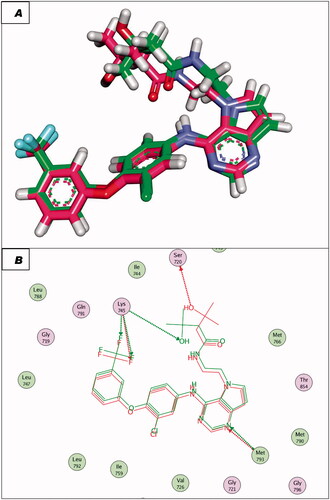
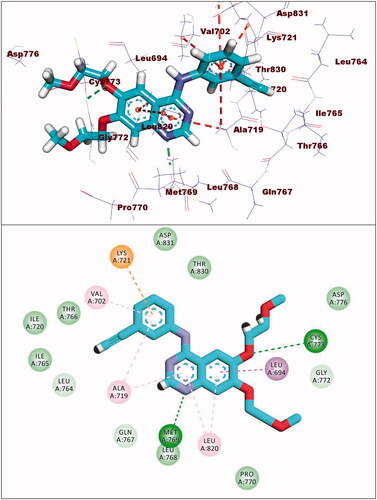
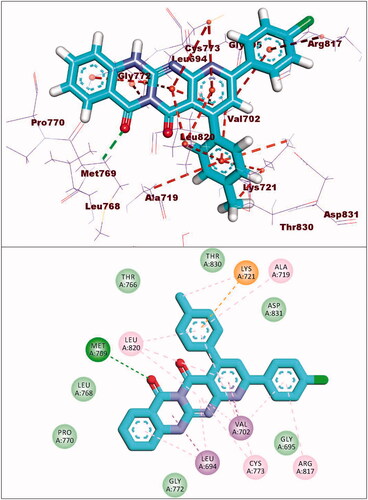
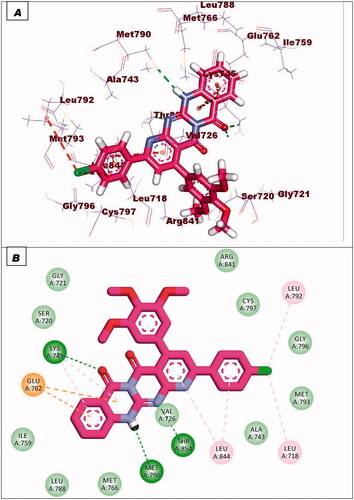
Figure 7. (A and B) 3D and 2D superimposition of the docked ligand (erlotinib; pink) and the original ligand (green) with RMSD value of 0.88 Å.
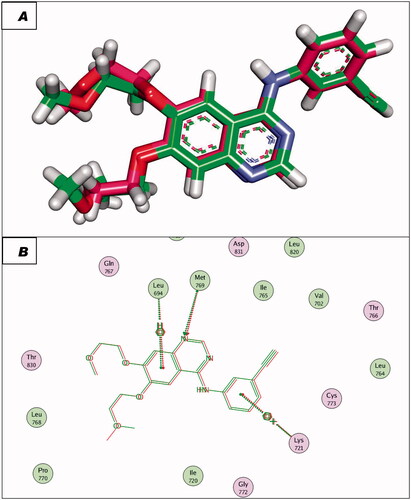
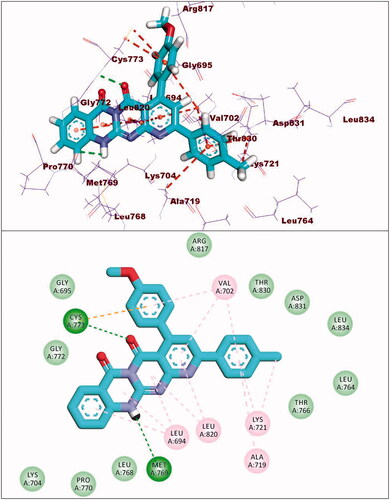
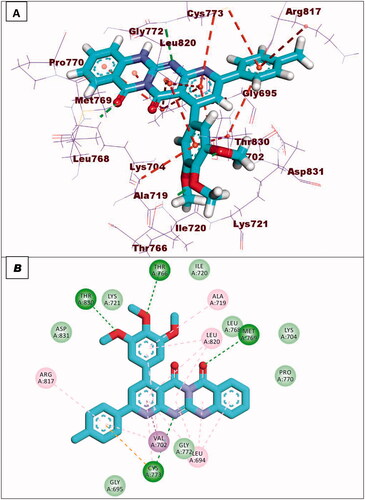
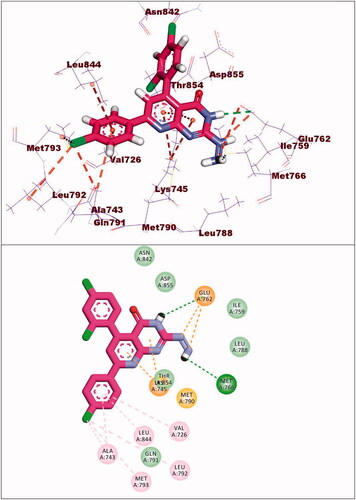
Figure 8. (A and B) 3D and 2D superimposition of the docked ligand of mutant EGFR (TAK-285; Pink) and the original ligand (green) with RMSD value of 1.06 Å.