Figures & data
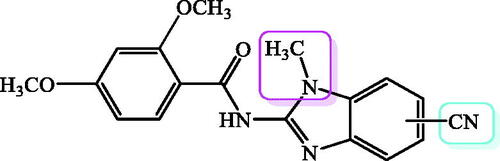
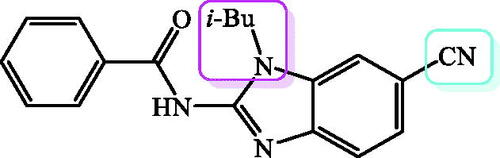
Figure 1. Recently published benzamides and benzimidazole-2-carboxamides with great antioxidative potential.
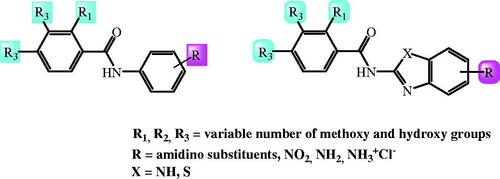
Table 1. IC50 values for 1,1-diphenyl-picrylhydrazyl (DPPH) free radical scavenging and ferricreducing/antioxidant power (FRAP) activities.
Table 2. Antiproliferative activity of tested compounds
Figure 4. Antioxidative activity of selected compounds in cells. HCT 116 cells were treated with the combination of tert-Butyl hydroperoxide (TBHP, 200 µM) and N-Acetyl-l-cysteine (NAC, 10 mM) or tested compounds 34 and 40 (10 µM) for 1 h. The level of reactive oxygen species (ROS) was measured with fluorescent dye 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) using fluorimeter. Treatment with 200 µM TBHP alone was used as a control for ROS induction. Data presented here are the results of two independent measurements, done in triplicates. One-way ANOVA with Tukey’s post hoc test was used for statistical analysis, *p < .05, **p < .01, ***p < .001.
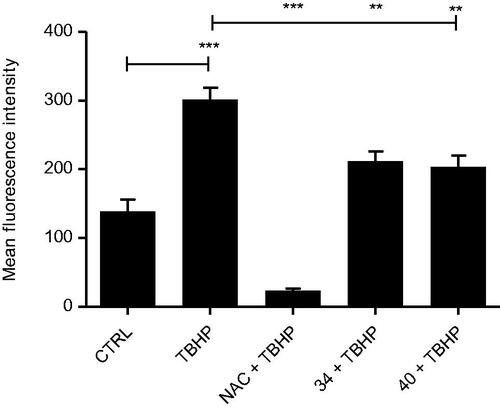
Figure 5. Nrf2 protein levels in the cytoplasmic and the nuclear fraction of HCT116 cells, assessed by Western blot analysis. HCT116 cells were treated with the 4 mM hydrogen peroxide as a positive control for ROS induction and with compounds 34 and 40 (50 µM and 5 µM, respectively) for 1 h. Representative western blots of Nrf2 in cytoplasm and the nuclear fraction are presented. Histone H3 and β-actin were employed as loading controls for nuclear and cytoplasmic fraction; respectively. Results are presented as mean ± SD; number of samples was n = 3.
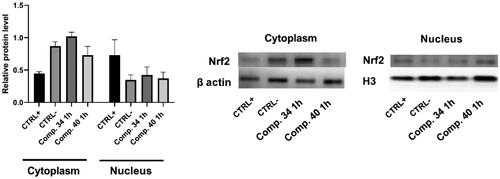
Figure 6. Products of the descriptor’s average value calculated for the dataset used to derive model and the associated PLS coefficient of 3D-QSAR model for: (A) model 1 and (B) model 2.
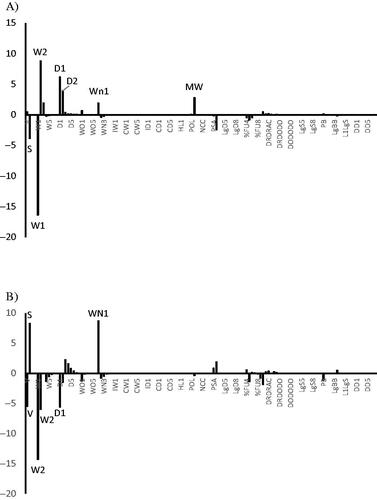
Table 3. Statistical properties of 3D-QSAR models.