Figures & data
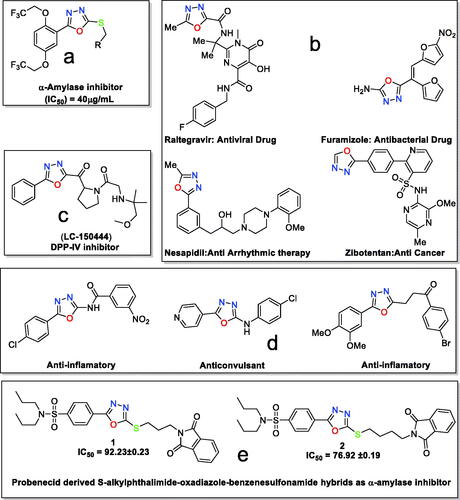
Figure 1. (a) 1,3,4-Oxadizole moiety containing compound as α-amylase inhibitor; (b) 1,3,4-oxadiazole moiety in drugs; (c) 1,3,4-oxadizole moiety containing compound in preclinical trials; (d) 1,3,4-oxdiazole displaying various biological activities; and (e) present work.
Scheme 1. Synthetic approach for probenecid derived three S-alkylphthalimide-oxadiazole-benzenesulfonamide hybrids (1 and 2).
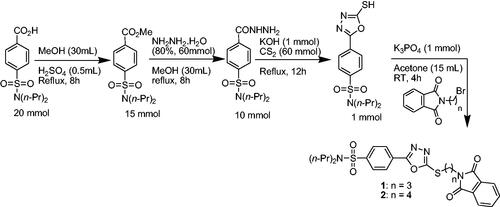
Figure 2. ORTEP diagram of hybrids (a) 1 and (b) 2 that are drawn at a probability level of 40%. Hydrogen atoms are shown by small circles of arbitrary radii. (c) Molecular overlay plot of hybrid 1, molecule I (red) and molecule II (blue). The major of the disordered propyl groups in hybrid 1 are shown for clarity.
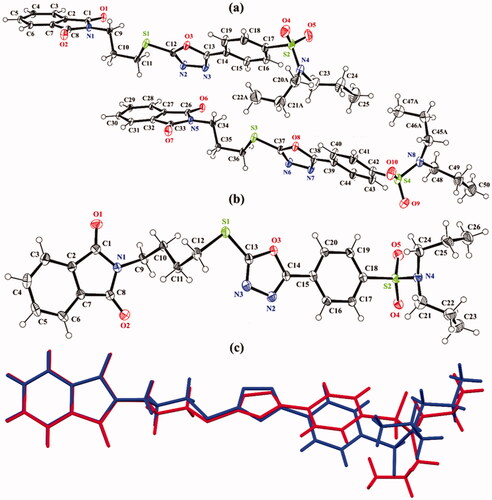
Table 1. X-ray Parameters of both hybrids 1 and 2.
Figure 3. Packing diagram of hybrids (a) 1, and (b) 2. Selected hydrogen atoms are shown for clarity. The major of the disordered propyl groups in hybrid 1 are shown for clarity.
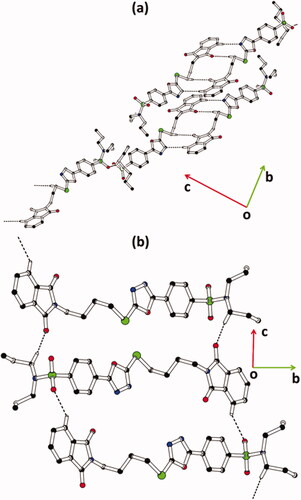
Figure 4. Offset ππ stacking interaction of hybrids (a) 1 and (b) 2. Hydrogen atoms are not shown. The major of the disordered propyl groups in hybrid 1 are shown for clarity. Sulphur atoms S1 and S3 are labelled in order to distinguish between molecules I and II of hybrid 1.
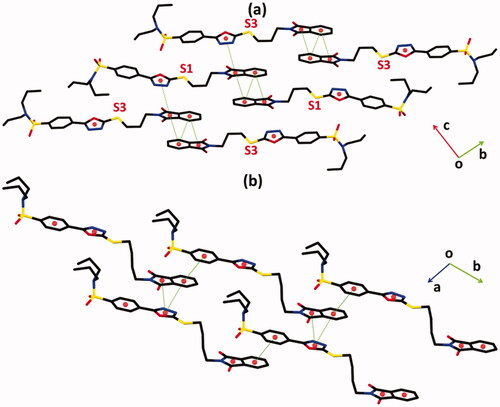
Table 2. Hydrogen-bond geometry (Å, °) for hybrids 1, 2.
Table 3. The important parameters of offset ππ stacking interactions in hybrids 1 and 2.
Figure 5. Molecular electrostatic potential (MEP) maps of hybrids (a) 1 and (b) 2 are plotted onto 0.002 au electron density contours. The electrostatic potential varies from −0.01 (red) to +0.01 (blue) au.
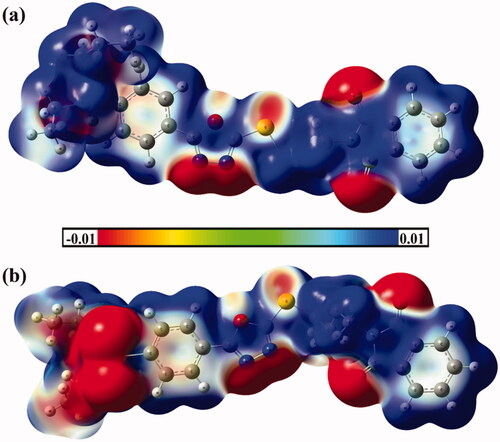
Figure 6. Frontier molecular orbitals (FMOs), including the highest occupied molecular orbitals (HOMO) and lowest unoccupied molecular orbitals (LUMO) of hybrids (a) 1 and (b) 2.
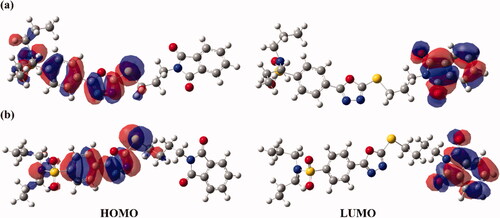
Figure 7. Quantum theory of atoms in molecules (QTAIM) and 3 D noncovalent interaction (NCI) isosurfaces of hybrids (a) 1 and (b) 2. The isosurfaces are generated with a reduced density gradient value of 0.50 au and coloured from blue to red according to sign(λ2)ρ ranging from −0.035 (blue) to 0.020 (red) au.
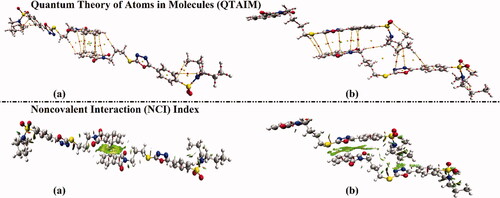
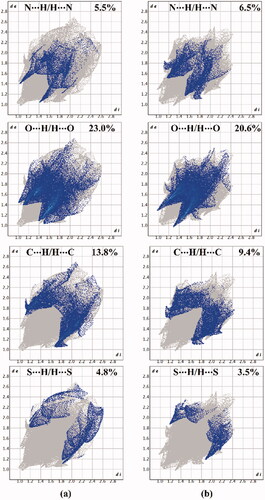
Figure 8. View of the Hirshfeld surfaces mapped over dnorm property of (a) hybrid 1 and (b) hybrid 2. The labels 1, 2, 3, and 4 represent N···H/H···N, O···H/H···O, C···H/H···C, and S···H/H···S interactions, respectively.
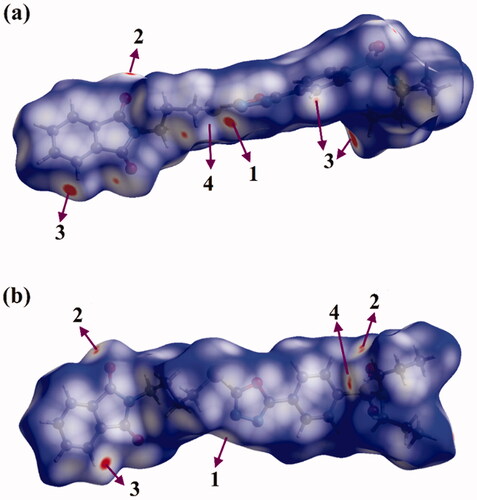
Figure 10. Hirshfeld surfaces of hybrids (a) 1 and (b) 2 mapped over Shape index and Curvedness properties.
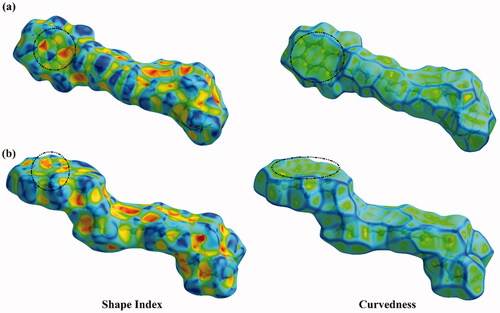
Figure 11. Graphical representation of α-amylase inhibition of probenecid derived two S-alkylphthalimide-oxadiazole-benzenesulfonamide hybrids (1 and 2) at different concentrations.
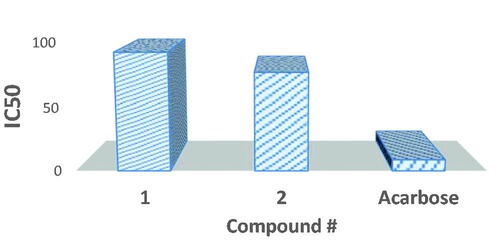
Table 4. α-Amylase inhibition values of probenecid derived two S-alkylphthalimide-oxadiazole-benzenesulfonamide hybrids (1 and 2).