Figures & data
Figure 2. Schematic presentation of pBVboostFG expression vector designed for production of recombinant protein. The insert contains: 1. attL1, 2. Shine-Dalgarno, 3. Kozak, 4. Met-Ser-Tyr-Tyr, 5. 6 x His, 6. Asp-Tyr-Asp-Ile-Pro-Thr-Thr, 7. Lys-Val, 8. β-CA gene of G. salaris gene of interest, 9. Two stop codons, and 10. AttL2.
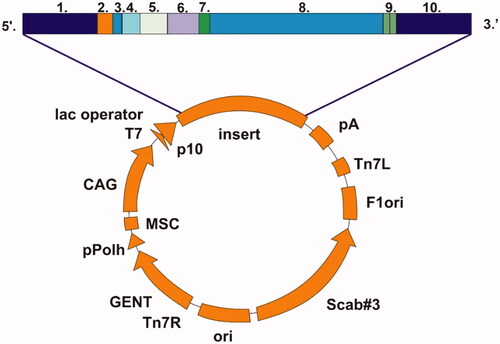
Figure 3. Alignment of GsaCAβ sequence with β-CA sequences of other metazoans. The conserved hallmark catalytic-site sequences of β-CAs, CXDXR and HXXC, are shown with black triangles (C: Cysteine, D: Aspartic acid, H: Histidine, R: Arginine, X: any residue). Columns with fully conserved residues are shown as red with white letters. Boxed columns denote positions in which at least 80% of residues are of similar type. The top line shows secondary structures derived from our GsaCAβ model. α: α-helices; β: β-strands; η: 310-helices; T: turns.
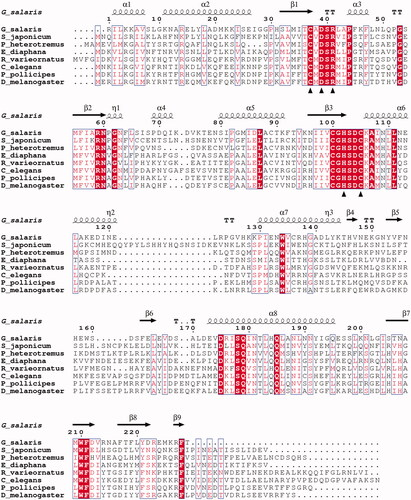
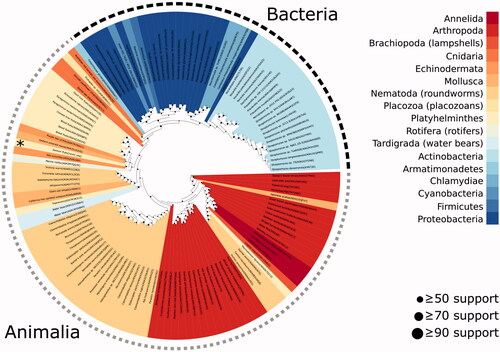
Table 1. Sequences in the multiple sequence alignment of .
Table 2. Percent identity matrix of the aligned protein sequences of , as computed by Clustal Omega
Figure 4. Recombinant GsaCAβ protein analysed on SDS-PAGE. The image shows protein ladder standards (left) and the purified recombinant G. salaris β-CA protein (right) showing a molecular mass calculated from mobility of 26.0 kDa.
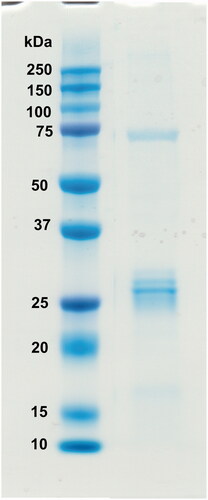
Table 3. Kinetic parameters for the CO2 hydration reactionCitation30 catalysed by α- and β-class CA enzymes: the human cytosolic isozymes hCA I and II (α-class CAs) at 20 °C and pH 7.5 in 10 mM HEPES buffer, Can2 (from Cryptococcus neoformans), CalCA (from Candida albicans), SceCA (from Saccharomyces cerevisiae), and Cab (from the archaeon Methanobacterium thermoautotrophicum) measured at 20 °C, pH 8.3 in 20 mM TRIS buffer and 10 mM NaClO4.
Table 4. Anion inhibition data of the β-CA from G. salaris and human isoforms hCA I and hCA II as determined by stopped-flow CO2 hydrase assay.
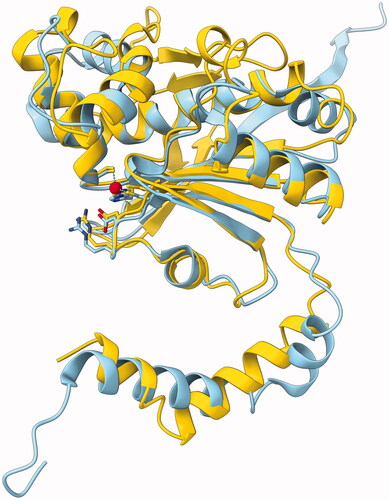
Figure 6. Molecular model of GsaCAβ. Our model constructed using AlphaFold, yellow, superimposed with pea β-CA (PDB 1EKJ, chain C), sky blue. The zinc ion of the catalytic site is shown in red.
Data availability statement
Data files pertaining to this study, including the 3 D protein model, are available at https://github.com/MarttiT/G.-salaris-BCA. Individual supplementary files in this repository are indicated as bit.ly links in the text. Further data files and code are stored at https://github.com/thirtysix/Aspatwar.Gsalaris_BCA.