Figures & data
Figure 1. FDA approved and reported kinases inhibitors with their essential pharmacophoric features.
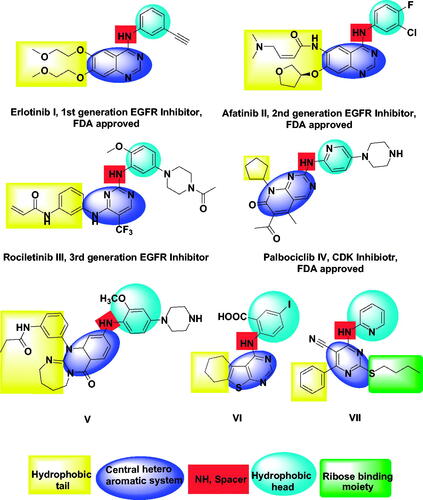
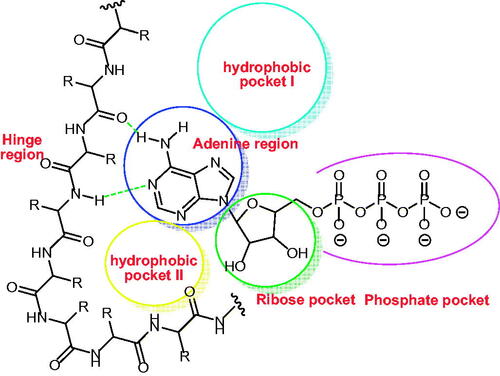
Figure 2. ATP binding site of EGFR cavity composed of five main partsCitation47–49.
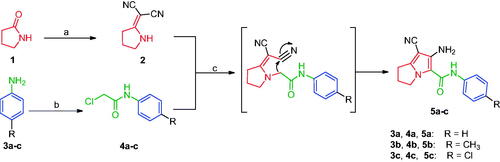
Scheme 1. Construction of compounds 5a–c. Reagents and conditions: (a) (CH3)2SO4, benzene, CH2(CN)2, reflux, 6 h; (b) ClCH2COCl, glacial acetic acid, CH2COONa, 30–40 °C, 2 h; (c) acetone, K2CO3, reflux, 24 h.
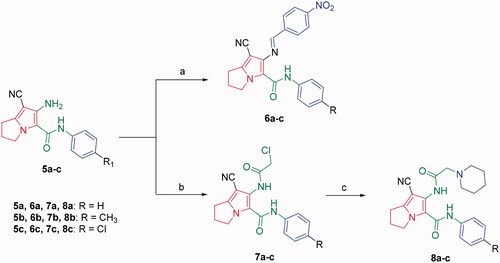
Scheme 2. Synthesis of compounds 6a–c, 7a–c and 8a–c. Reagents and conditions: (a) 4-Nitrobenzaldehyde, absolute ethanol, glacial acetic acid; (b) ClCH2COCl, benzene; r.t.; 48 h; (c) piperidine, NaHCO3, absolute ethanol, reflux, 8 h.
Scheme 3. Synthesis of compounds 9a–c, 10a–c and 11a–c. Reagents and conditions: (a) Excess CH3C(OC2H5)3, reflux, 12 h; (b) K2CO3, DMF, r.t., 48 h; (c) ClCH2COOC2H5, K2CO3, acetone, reflux, 6 h.
Table 1. IC50 values of the new compounds (6a–c, 8a–c, 9a–c, and 11a–c) against Hep3B, HCT116 and MCF-7 cell lines.
Table 2. Inhibitory activity of compound 8b and imatinib against 20 kinases at 10 µMCitation66.
Figure 4. Cell cycle distribution of MCF-7 treated with compound 6a (µM, 72 h: x axis); % cell (y axis).
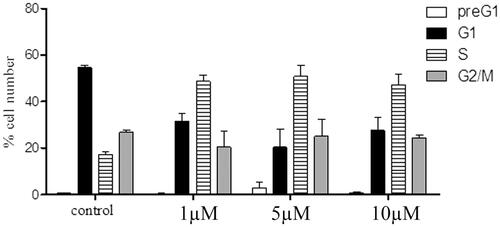
Figure 5. Cell cycle distribution of MCF-7 treated with compound 8b (µM, 72 h: x axis); % cell (y axis).
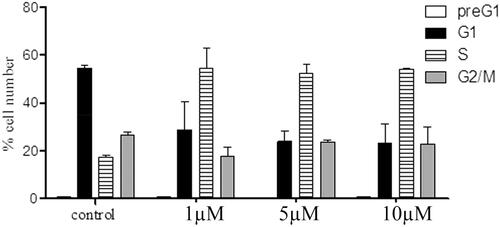
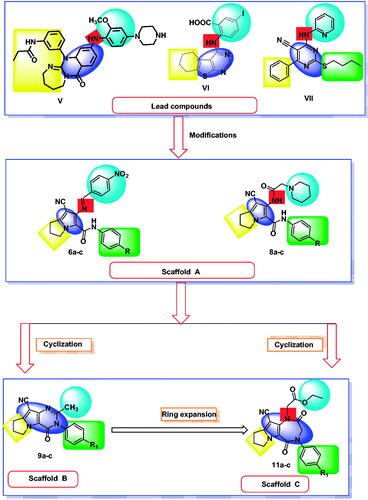
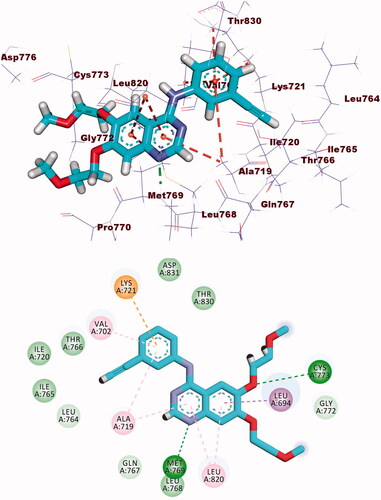
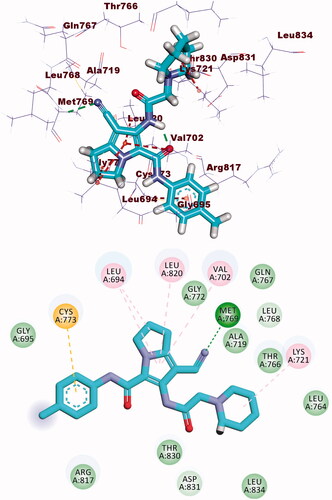
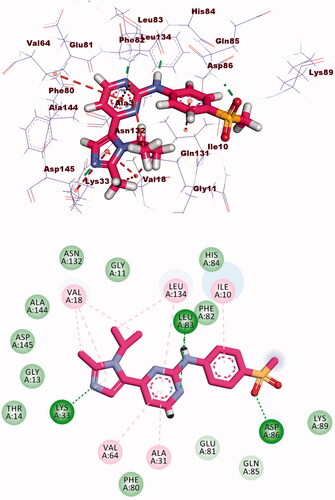
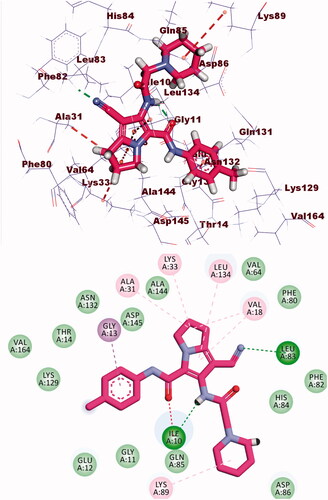
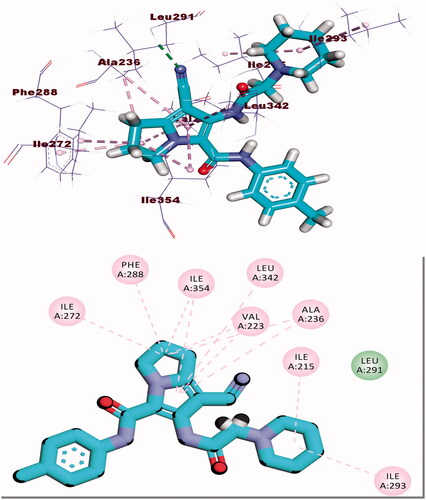
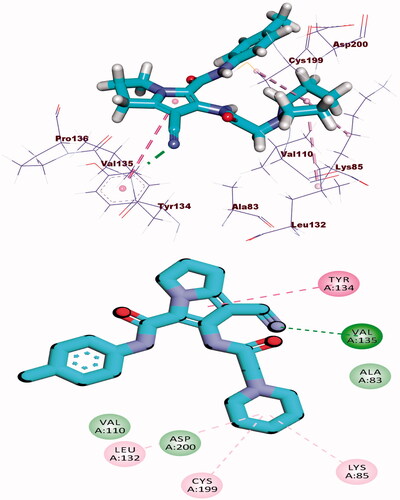
Table 3. The binding free energies of the synthesised compounds against EGFR and CDK-2.
Figure 6. (A) superimposition of the docked ligand of erlotinib (turquoise) and the original ligand (green) with an RMSD value of 0.88 Å. (B) superimposition of the docked ligand of AZD5438 (pink) and the original ligand (green) with RMSD value of 0.54 Å.
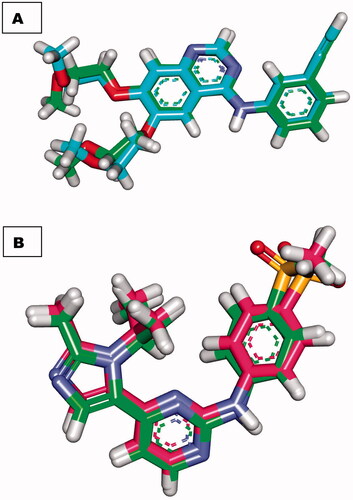
Table 4. Toxicity properties of the synthesised compounds.
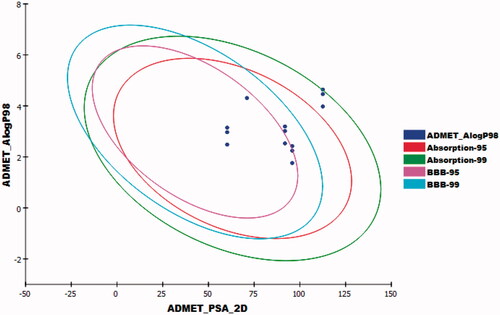
Table 5. Predicted ADMET profile for the synthesised compounds