Figures & data
Figure 1. (a) Inactive conformation of c-Src shows phosphorylated Tyr530 on the SH2 domain; (b) Phosphorylation of Tyr419 on the kinase domain SH1 allows to activate of the enzyme.
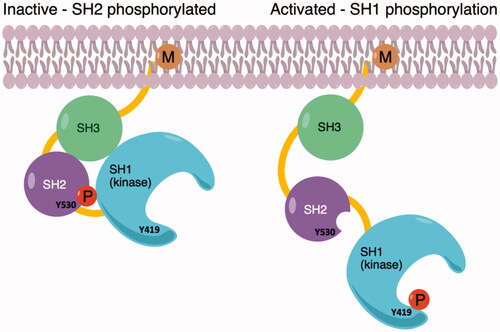
Figure 2. Structures of the molecules approved by the FDA or in clinical trials as Src-family inhibitors.
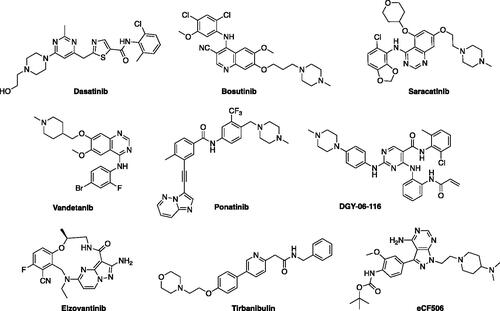
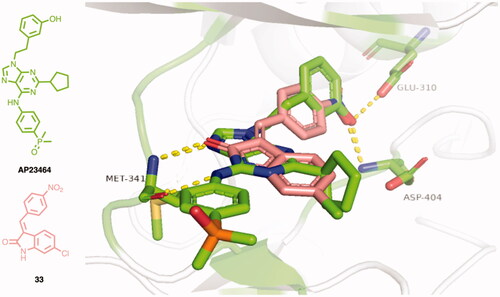
Table 1. Evaluation of the inhibitory activity of c-Src by the in-house prepared collection of small molecules.
Table 2. Inhibitory activity evaluation of indolinones 22–30.
Scheme 1. General synthesis by Knoevenagel condensation starting from 6-chloro oxindole and different aromatic and heteroaromatic compounds.
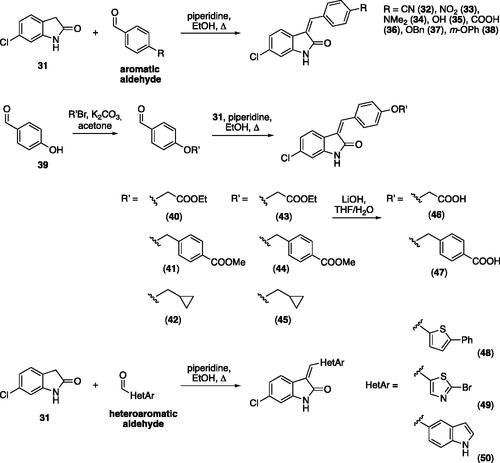
Table 3. Evaluation of the inhibitory activity of the novel indolinone derivatives.
Table 4. Cytotoxicity evaluation of selected compounds on human MCF-7 breast cancer cell line.