Figures & data
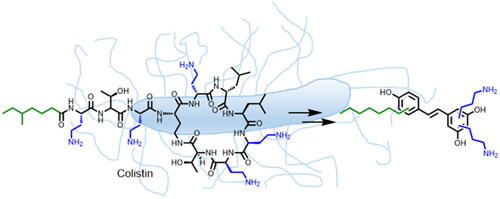
Figure 1. Colistin inspiration for aminoalkyl resveratrol derivatives (resveratrol, RES drawn in black colour.
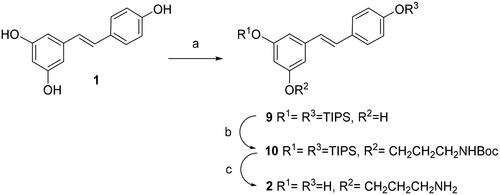
Scheme 1. Synthesis of compound 2. Reagents and conditions: (a) TIPS chloride (2.0 equiv), DIPEA (2.0 equiv), DMAP (2.0 equiv), DMF, -15° C for 10 min, then rt for 16 h; (b) ICH2CH2CH2NHBoc, K2CO3, DMF; (c)TFA, THF-H2O.
Scheme 2. Synthesis of alkyl resveratrol derivatives 11–14. Reagents and conditions: (a) 1-iodobutane, 1-bromooctane or 1-bromodecane, K2CO3, DMF.
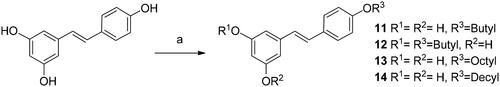
Scheme 3. Synthesis of monoamino alkyl resveratrol derivatives 3–4. Reagents and conditions: (a) ICH2CH2CH2NHBoc or BrCH2CH2NHBoc, K2CO3, DMF; (b) TFA, THF.
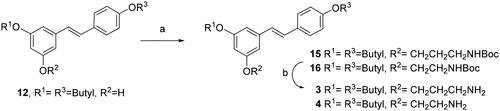
Scheme 4. Synthesis of diamino and triamino alkyl resveratrol derivatives 5–8. Reagents and conditions: (a) i. BrCH2CH2CH2Cl, K2CO3, DMF, 80 °C, 5 h. ii) NaN3, DMF, 50 °C, 16 h; (b) PPh3, THF, 16 h.
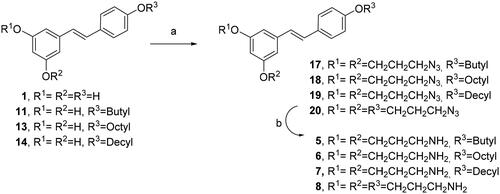
Table 1. Minimal inhibition concentration (MIC) observed for the different RES derivatives against a panel of Gram-negative and Gram-positive aerobically growing bacteria. -, these bacteria were resistant to the higher tested concentration (128 µM).
Table 2. Minimal inhibition concentration (MIC) observed for the different RES derivatives against a panel of Gram-negative and Gram-positive anaerobic bacteria. -, these bacteria were resistant to the higher tested concentration (128 µM).
Figure 3. MICs distribution according to the hydrophobicity (LogP) of the amino RES derivatives. A) Gram-negative aerobic bacteria, B) Gram-positive aerobic bacteria, C) Gram-negative anaerobic bacteria, D) Gram-positive anaerobic bacteria. Each dot is representing the MIC for a tested bacteria against each compound. For those bacteria for which MIC was not reached, 256 µM was considered for this graph.
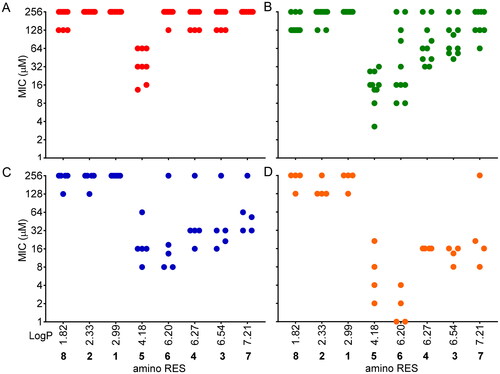
Figure 4. Activity of amino RES derivatives on the outer membrane of E. coli LMG8224. A) Outer-membrane permeabilization by different amino RES 5 concentrations (µM). RES 32, resveratrol control at 34 µM, PolB polymyxin B positive control at 4 µM. Neg, negative control. RFU, random fluorescence units. Effect of the outer membrane main component LPS (B) and the outer membrane stabilising agents Mg2+ (C) and Ca2+ (D) on the antimicrobial activity of amino RES 5. For the points labelled with *, the MIC was higher than 128 µM.
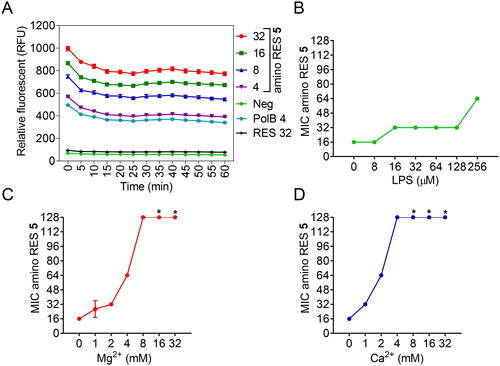
Figure 5. Membrane permeabilization by amino RES 5 in E. coli LMG 8224 (A) and Gram-positive bacteria B. cereus ATCC 10987 (B). RFU, random fluorescence units. C, membrane depolarisation (DiSC3(5)) for E. coli LMG 8224 and B. cereus ATCC 10987 (D). RES, resveratrol control, Pol B positive control and Gra, gramicidin S positive control. The concentrations are expressed in µM. E and F, bactericidal mode of action for E. coli LMG 8224 and B. cereus ATCC 10987 respectively. The data are expressed as % of the cell growth respect the negative control.
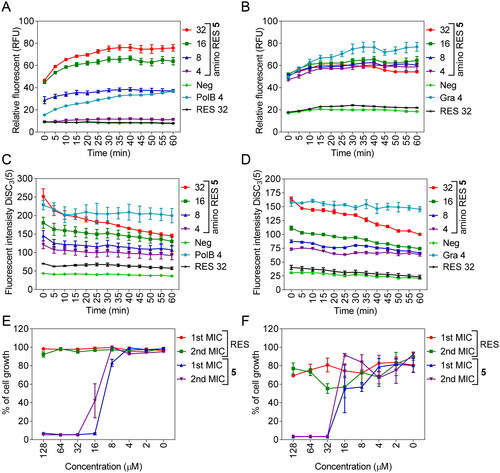
Table 3. Combined activity of amino RES 5 and traditional antibiotics against selected Gram-negative bacteria. AB, A. baumannii LMG 01041, ECl, E. cloacae LMG 02783. ECo, E. coli LMG 8224, KE, K. aerogenes LMG 02094, KP, K. pneumoniae LMG 20218, SE, S. enterica LMG 07233.
Table 4. Haemolityc activity against human red cells and cytotoxicity of the amino RES derivatives. The concentrations are expressed in μM ± SE.