Figures & data
Table 1. Fragment library subsets and number of fragments used for screening on DdlB.
Table 2. Profiling of repurchased fragment hits from the screening.
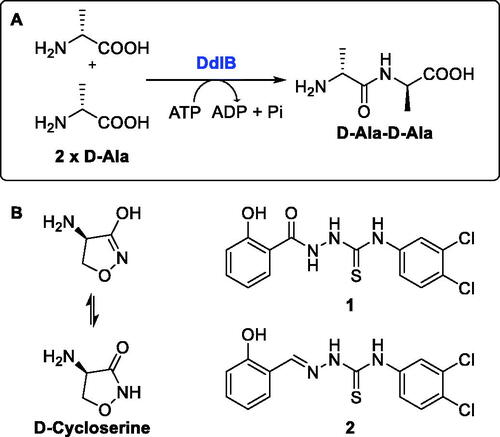
Figure 2. Fragment hit 3 targeting DdlB. (A) Dose–response inhibition of DdlB for 3. (B) SPR analysis of 3 with the immobilised DdlB. 3 was injected across DdlB in serial dilutions ranging from 78.125 to 5000 µM for 40 s at a flow rate of 30 μL/min, and dissociation was followed for 40 s. Three independent measurements were performed and representative sensorgrams revealing dose-dependent binding of 3 to DdlB are shown. (C) Data were fitted to the steady-state affinity model to obtain the apparent equilibrium dissociation constant (KD). KD values are the mean ± standard deviation of three titrations. (D) ATP (blue sticks) and D-Ala-D-Ala (white sticks) binding sites of DdlB (PDB ID: 4C5C) with the co-crystallised ligands. (E) Computational docking of 3 to the ATP-binding site using extra-precision Glide (blue sticks) and QM-Polarized Ligand Docking (orange sticks) (PDB ID: 4C5C).
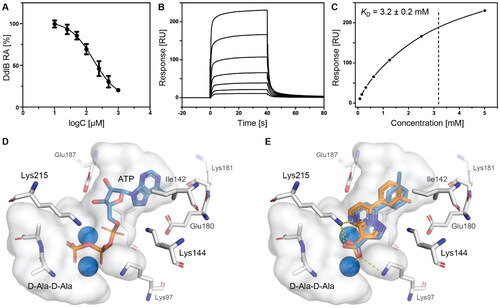
Table 3. Analogues of compound 3 that were evaluated in the DdlB inhibition assay.
Table 4. The breakdown of GlideScore scores into individual contributions, only non-zero terms are shown.