Figures & data
Figure 2. Compound 5f as a representative example of the synthesised compounds occupying the ATP active cavity of EGFR (schematic presentation).
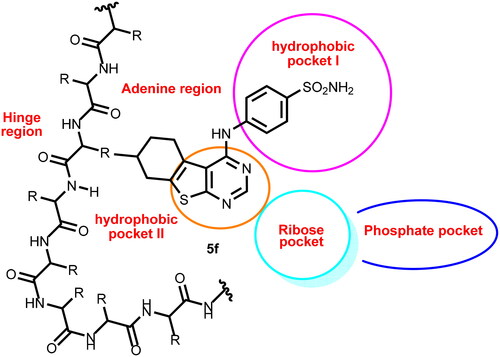
Table 1. In vitro anti-proliferative and EGFR inhibitory activities of the tested compounds.
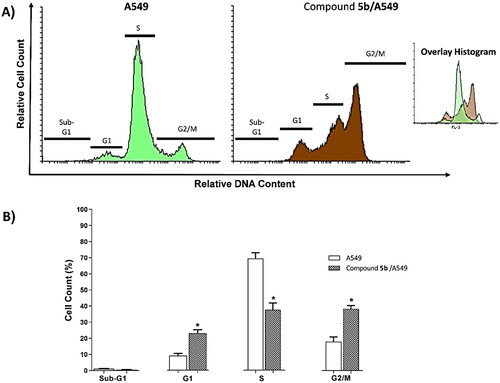
Table 2. Effect of 5b on cell cycle progression in A549 cells after 72 h treatment.
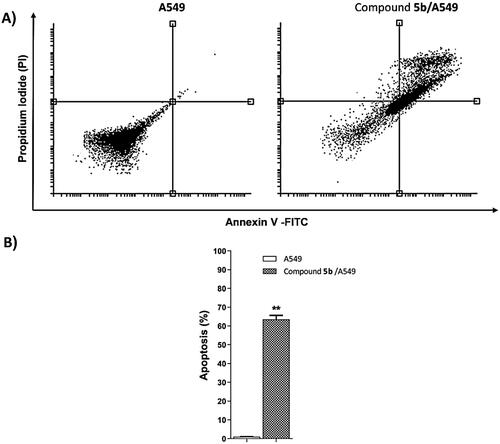
Table 3. Effect of compound 5b on stages of the cell death process in A549 cells after 72 h treatment.
Figure 6. Gene expression analysis of BAX and Bcl-2 the expression levels after treatment of A549 with compound 5b for 72h.
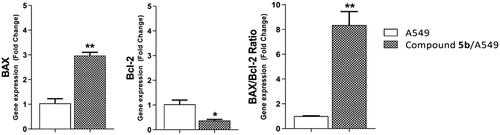
Table 4. Effect of 5b on levels of BAX and Bcl-2 genes expression in A549 cells treated for 72 h.
Table 5. The docking binding free energies of the synthesised compounds against EGFRWT and EGFRT790M.
Figure 7. Erlotinib docked into the active site of EGFRWT forming two HBs with Met769 and Cys773 and seven HIs with Lys721, Val702, Ala719, Leu820, and Leu694.
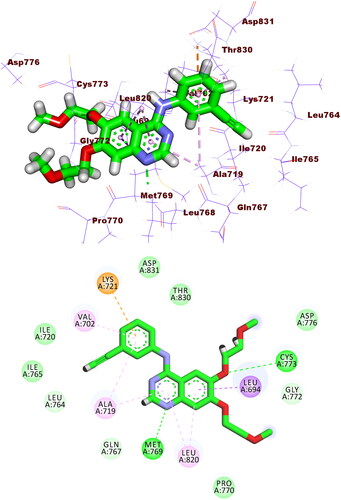
Figure 8. Compound 5b docked into the active site of EGFRWT forming 1 HB with Met769 and 12 HIs with Lys721, Val702, Ala719, Leu820, Cys773, and Leu694.
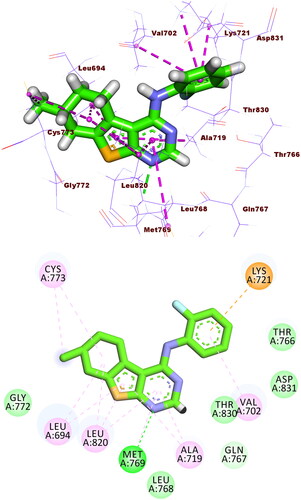
Figure 9. Compound 5f docked into the active site of EGFRWT, forming 1 HB with Met769, and 10 HIs with Val702, Ala719, Leu820, Cys773, and Leu694.
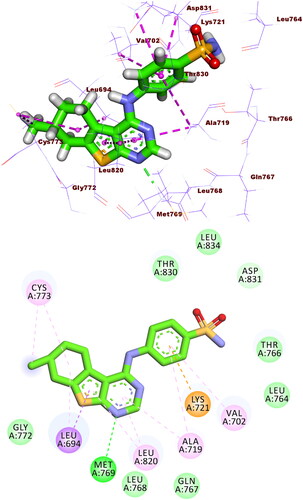
Figure 10. Co-crystallised ligand (TAK-285) docked into the active site of EGFRT790M forming formed 3 HBs with Met793, Ser720, and Lys745 and 14 HIs with Lys745, Glu762, Leu788, Ile759, Leu844, Ala743, Val726, Met790, and Ala743.
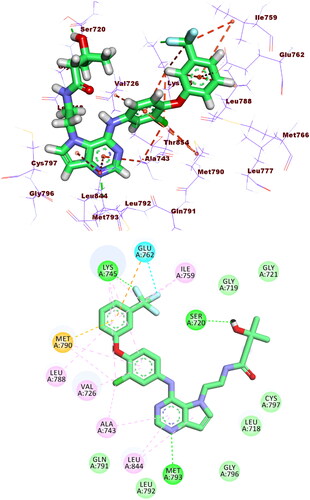
Figure 11. Binding of compound 5f with EGFRT790M, forming two HBs with Met793 and Lys745, nine HIs with Met766, Leu788, Ile759, Val726, Met790, Ala763, and Ala743, and three electrostatic attractions with Met790, Lys745, and Glu762.
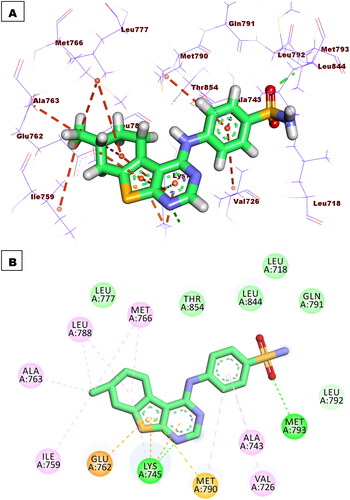
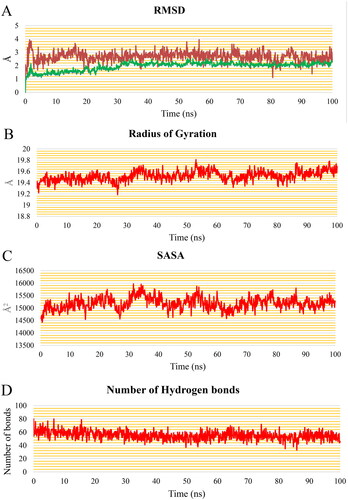
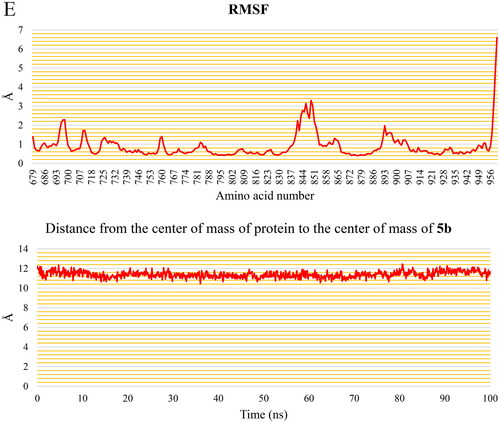
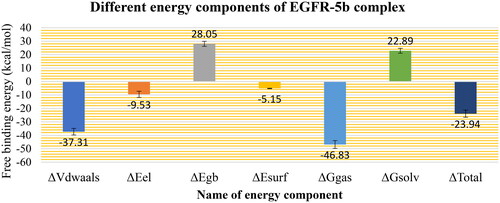
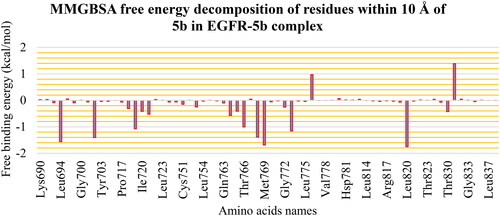
Figure 15. PLIP analysis of the EGFR-5b complex for each cluster representative. HB: blue solid line, HI: dashed grey line, amino acids: blue sticks, and 5b: orange sticks.
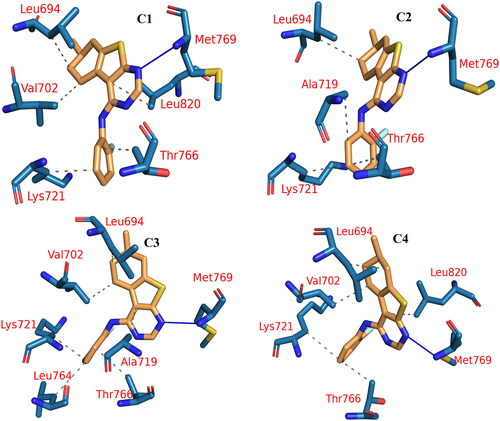
Table 6. The number and types of interactions detected from the PLIP webserver.
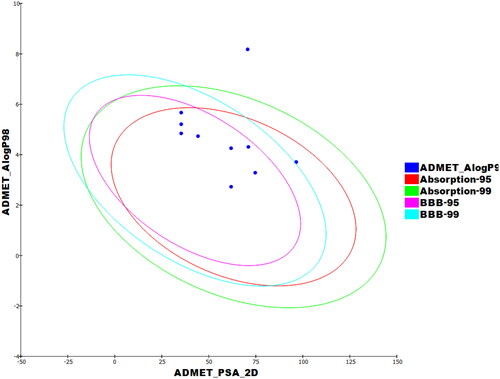
Table 7. Calculated ADMET descriptors.
Table 8. Toxicity properties of the synthesised compounds.