Figures & data
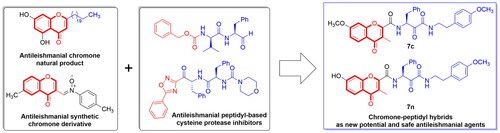
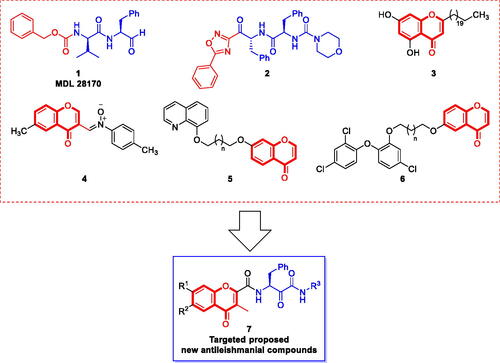
Figure 1. Chemical structures of cysteine protease inhibitors with potential antileishmanial activity and the design rational of chromone-peptidyl hybrids as new antileishmanial agents.
Scheme 1. Synthesis of target chromone-peptidyl hybrids (7a–q). Reagents and conditions: (i) EDC, HOBt, DMF, stir 0 °C-rt, 2h; (ii) Dess-Martin periodinane, DMF, 0 °C-rt, 10% Na2S2O3 solution, 2h (7a–l); (iii) 1% methanolic HCl, 70 °C, 2 h (7m–q).
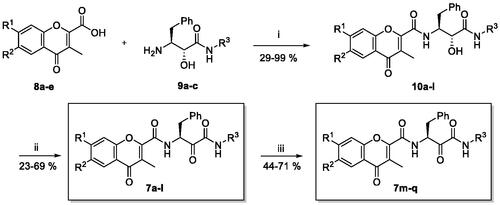
Table 1. % Growth inhibition of L. donovani promastigotes elicited by chromone peptidyl hybrids (7a–q).
Table 2. Determined IC50 values against L. donovani promastigotes for the most active chromone peptidyl hybrids.
Table 3. In vitro evaluation of cytotoxicity on THP-1 cell of compounds 7c, 7h, and 7n.
Figure 2. Sequence alignment and Ramachandran plot of the generated homology model of CALP of L. donovani (A) Sequence alignment against calpain I protease core (PDB code: 1TL9); (B) Ramachandran plot of amino acid residues of the generated homology model indicated that most of them are in the favoured regions.
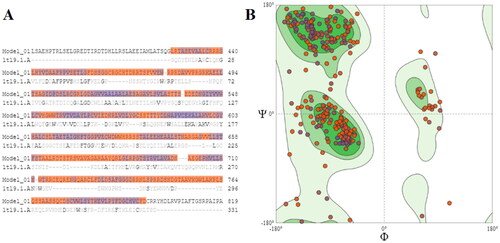
Figure 3. Position of active site in three-dimensional protein structure showing co-crystallized inhibitor leupeptin in the active site.
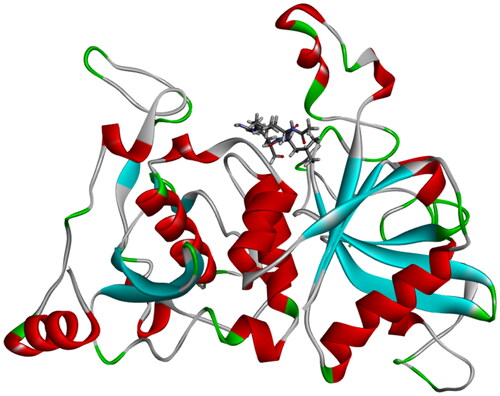
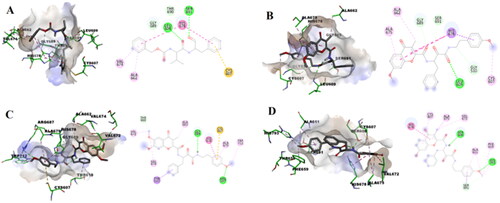
Figure 4. Predicted binding modes of compounds 7c, 7h, and 7n as well as the reference ligand MDL28170 with LdCALP: (A) Calculated docking pose of the reference compound MDL28170 (1) in 3D (left) and 2D view (right) into the substrate pocket of LdCALP; (B) Calculated docking pose of compound 7c in 3D (left) and 2D view (right) into the substrate pocket of LdCALP; (C) Calculated docking pose of compound 7h in 3D (left) and 2D view (right) into the substrate pocket of LdCALP; (D) Calculated docking pose of compound 7n in 3D (left) and 2D view (right) into the substrate pocket of LdCALP.