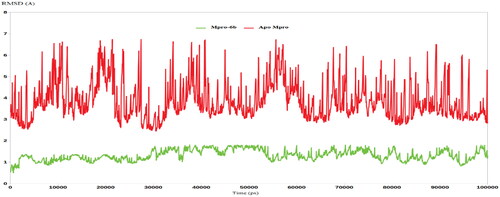
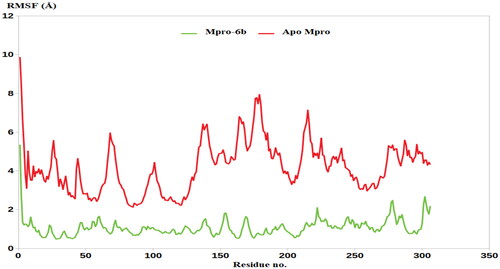
Figures & data
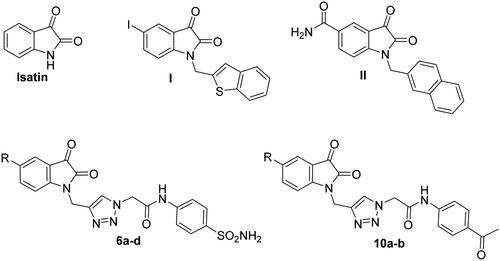
Figure 1. Structures of isatin and some reported main SARS-CoV protease inhibitors (I and II), as well as the target triazolo isatins (6a-d and 10a-b).
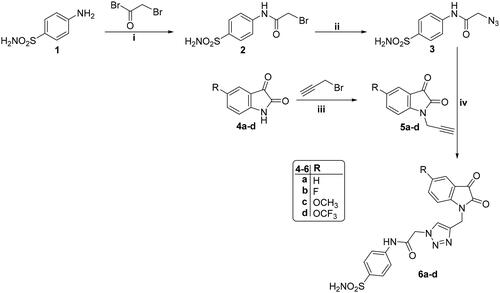
Scheme 1. Reagent and conditions: (i) Dry dioxane, K2CO3, stirring r.t., 12 h; (ii) NaN3, DMF, stirring r.t., 8 h; (iii) Dry acetonitrile, K2CO3, Stirring r.t., 10 h; (iv) DMF/H2O, CuSO4.5H2O, sodium ascorbate, heating at 60 °C, 7 h.
Scheme 2. Reagent and conditions: (i) Dry dioxane, K2CO3, stirring r.t., 12 h; (ii) NaN3, DMF, stirring r.t., 8 h; (iii) 5a or 5c, DMF/H2O, CuSO4.5H2O, sodium ascorbate, heating at 60 °C, 7 h.
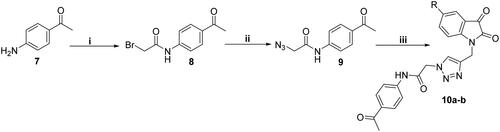
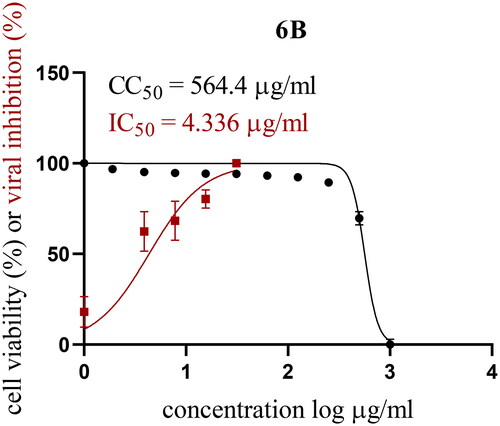
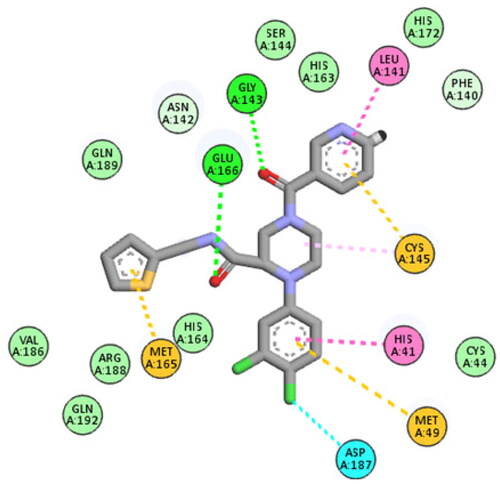
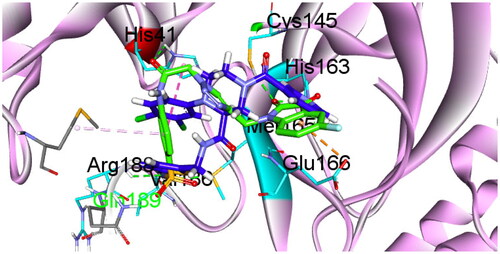
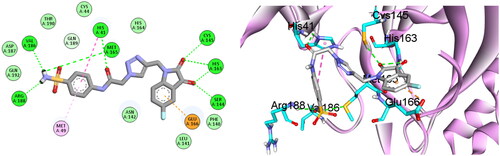
Table 1. In vitro inhibitory effect of target triazolo isatins (6a-d and 10a-b) against 3CL-Pro, using (GC376) as a standard drug.