Figures & data
Figure 2. Structures of some reported pyrimidine or thiouracil-based derivatives (VII, VIII, and IXa–b) as potent anti-mycobacterial agents.
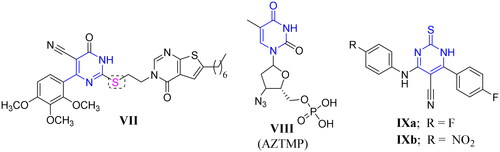
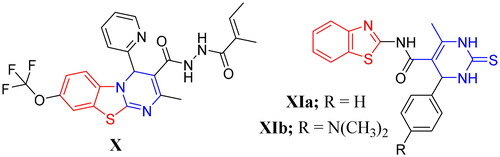
Figure 3. Examples of some benzothiazole derivatives (X and XIa–b) incorporating pyrimidine moiety as potent anti-mycobacterial agents.
Scheme 1. Synthesis of compounds 2–4; conditions and reagents: (a) dry benzene, anhydrous K2CO3, reflux for 12 h; (b) absolute ethyl alcohol, anhydrous K2CO3, reflux for 10–12 h; (c) dry acetone, anhydrous K2CO3, reflux for 8–10 h.
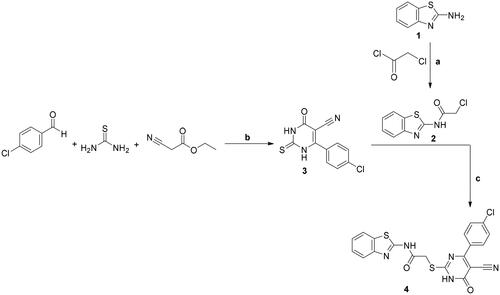
Scheme 2. Synthesis of 5a–c and 6; conditions and reagents: (a) dry DMF, CH3I/C6H5CH2Cl/ClCH2COOC2H5, anhydrous K2CO3, reflux for 12 h; (b) POCl3, reflux for 3 h.
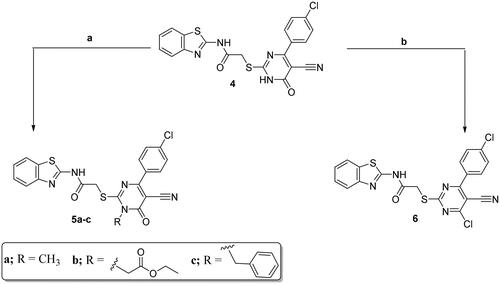
Scheme 3. Synthesis of compounds 7a–f and 8–13; conditions and reagents: (a) absolute ethyl alcohol, primary or secondary amine, TEA, room temperature for 24 h, then the heating under reflux for 6–12 h; (b) hydrazine hydrate, abs. ethanol, reflux, 6 h; (c) thiourea, abs. ethanol, reflux, 6 h; (d) n-butanol, glycine, reflux for 3 h; (e) reflux for 2 h with acetic anhydride; (f) fusion with the anthranilic acid in the oil bath at 190 °C, for 2 h; (g) glacial acetic acid, sodium azide, reflux for 3 h.
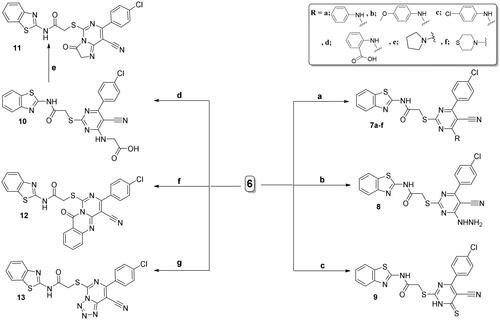
Scheme 4. Synthesis of 14 and 15; conditions and reagents: (a) glacial acetic acid and acetyl acetone, reflux for 6 h. (b) Reflux with the ethyl acetoacetate in NaOC2H5, for 4 h.
Table 1. MIC in µg/mL for 4, 5a–c, 6, 7a–f, and 8–15, as well as INH against the sensitive M. tuberculosis (ATCC 25177).
Table 2. MIC in µg/mL of 4, 5a–c, 6, 7e, 7f, 8, 9, 12, 14, and 15 against the MDR TB (ATCC 35822) and XDR TB (RCMB 2674) strains.
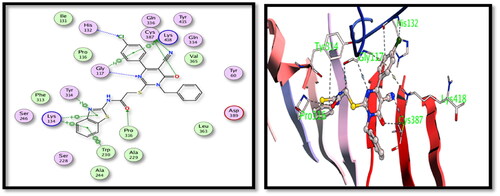
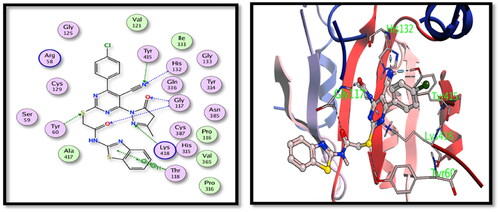
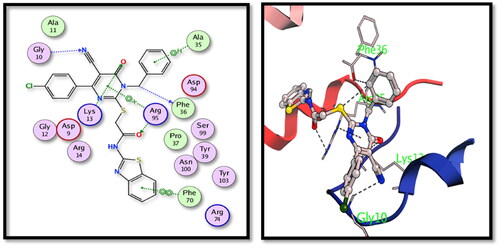
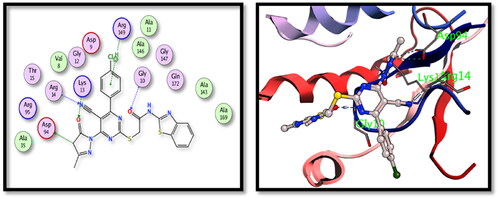
Table 3. Binding energy results in kcal/mol for the tested hybrids versus the co-crystallised ligands.