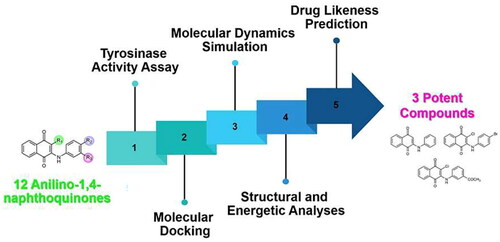
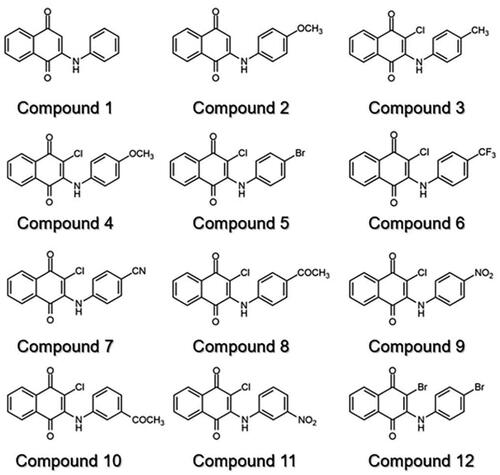
Figures & data
Figure 1. (A) Crystal structure of tyrosinase (PDB ID: 2Y9XCitation2). The catalytic histidine residues and copper ions (CuA and CuB) were labelled in purple and orange, respectively. (B) Melanogenesis pathway produces eumelanin and pheomelaninCitation3. (C) Chemical structures of KA, lawsone, plumbagin, and 2-phenyl-1,4-naphthoquinones.
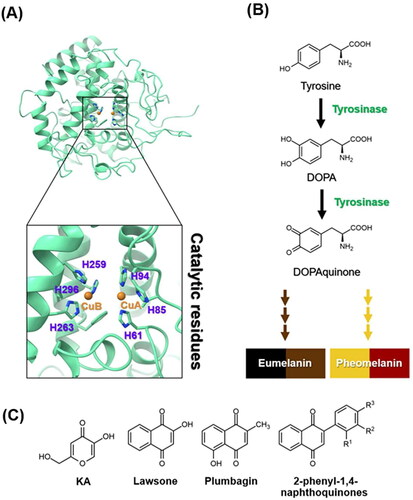
Figure 3. Mushroom tyrosinase inhibitory activity of 12 anilino-1,4-naphthoquinone derivatives at a concentration of 160 μM. Data are shown as the mean ± standard error of the mean (SEM) (n = 3). **p < 0.01, ***p < 0.001, and ****p < 0.0001 vs. control.
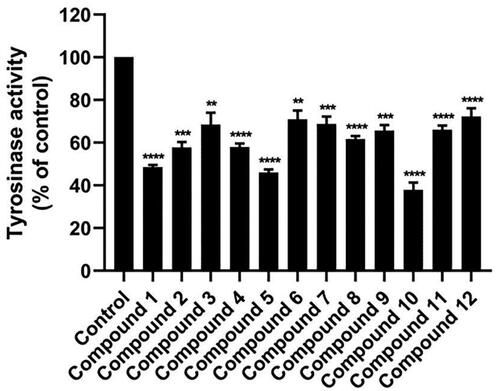
Figure 4. Tyrosinase inhibitory activity of (A) compound 1, (B) compound 5, (C) compound 10, and (D) KA using l-DOPA as a substrate. Data are shown as the mean ± SEM (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 vs. control.
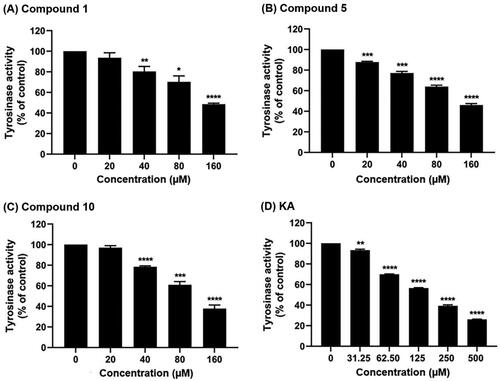
Table 1. IC50 values of compounds 1, 5, 10, and KA against tyrosinase.
Figure 6. Time evolution of (A) RMSD and (B) Rg of compounds 1, 5, 10, and KA in complexes with tyrosinase.
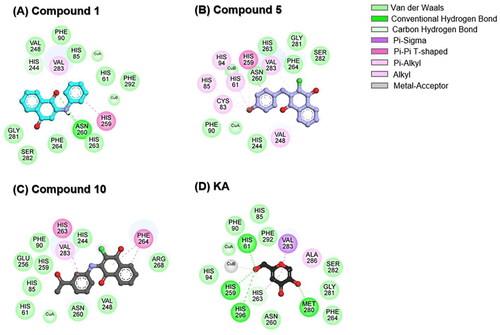
Table 2. CB-DOCK2 interaction energy of compounds 1, 5, 10, and KA against tyrosinase.
Figure 5. 2D interaction profile of (A) compound 1, (B) compound 5, (C) compound 10, and (D) KA in complexes with tyrosinase.
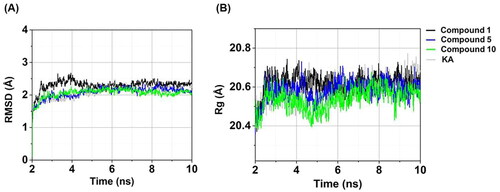
Figure 7. Time evolution of (A) #Contacts and (B) SASA of compounds 1, 5, 10, and KA in complexes with tyrosinase.
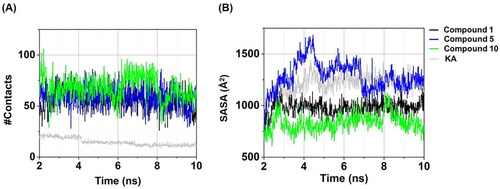
Figure 8. (Left) ΔGbind, res of compounds 1, 5, 10, and KA in complexes with tyrosinase. (Right) Representative structures showing the ligand orientation in the catalytic site drawn from the last 5 ns MD snapshots. The copper ions were hidden. The residues involved in the ligand binding (energy stabilisation of ≤ −1.0 kcal/mol) were coloured according to their ΔGbind, res values, where the highest to lowest ΔGbind, res values were shaded from yellow to red, respectively.
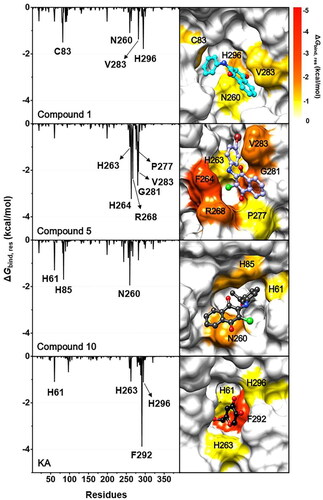
Table 3. Predicted values of drug-likeness parameters according to Lipinski’s rule of five criteria for compounds 1, 5, 10, and KA.
Data availability statement
The datasets presented in the current study are available upon reasonable request.