Figures & data
Figure 1. Genome sequencing costs as compiled by the National Human Genome Research Institute since 2001. In this figure, adapted from data provided by the National Human Genome Research Institute [Citation15], the cost of sequencing a single human genome is shown to drop precipitously by 100,000-fold over less than 20 years. This precipitous drop in the cost of sequencing has made it feasible to compare the entire genome of a single cancer patient to their own normal (noncancerous) genome, on a routine basis.
![Figure 1. Genome sequencing costs as compiled by the National Human Genome Research Institute since 2001. In this figure, adapted from data provided by the National Human Genome Research Institute [Citation15], the cost of sequencing a single human genome is shown to drop precipitously by 100,000-fold over less than 20 years. This precipitous drop in the cost of sequencing has made it feasible to compare the entire genome of a single cancer patient to their own normal (noncancerous) genome, on a routine basis.](/cms/asset/85ddec73-22f4-40ea-af0a-3543d5a79281/ierv_a_2012456_f0001_b.gif)
Figure 2. The Ancer pipeline. Tumor/normal Next Generation Sequencing (NGS) data is inputted in Ancer which first identify somatic variants and HLA types of the patient. Mutations are then processed with EpiMatrix to identify ones that generate HLA class I or HLA class II neoepitopes. Neoepitopes are then refined with JanusMatrix to remove sequences capable of eliciting regulatory T cells or other deleterious cross-reactive T cells. Neoantigens containing multiple neoepitopes are then automatically designed by the platform and subsequently ranked using proprietary parameters. The Ancer pipeline is a locked process aimed at minimizing human and manual interventions.
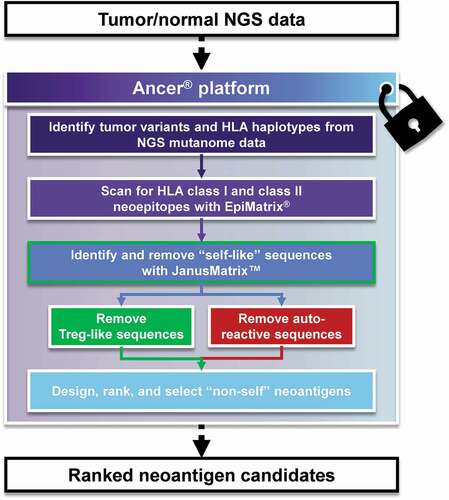
Figure 3. Neoantigen vaccine administration timeline. Patients have for the most part received NEO-PCVs in adjuvant settings due to slow design and production. Optimized vaccine design systems that employ diagnostic biopsies to identify neoantigens along with accelerated manufacturing and formulation processes allow vaccines to be administered in the neoadjuvant setting, where patients and their immune system are overall healthier. CPI: checkpoint inhibitor.