Figures & data
Figure 1. Overview of the involvement of muscle stem cells in regenerative myogenesis. The color of the transcription factors marks their presence (green) versus absence (red) in specific cell types. FAPs, fibro-adipogenic progenitors; MRFs, myogenic regulatory factors; MSCs, mesenchymal stem cells/multipotent stromal cells; MYOD1, myoblast determination protein 1; MyHC, myosin heavy chain; MYOG, muscle-specific transcription factor myogenin; PAX7, paired box 7; SCs, satellite cells.
Figure 2. Bioinformatic STRING analysis of major myosin components in the sarcomere of mouse skeletal muscles. MyHC, myosin heavy chain; MLC, myosin light chain; MYBP, myosin-binding protein.
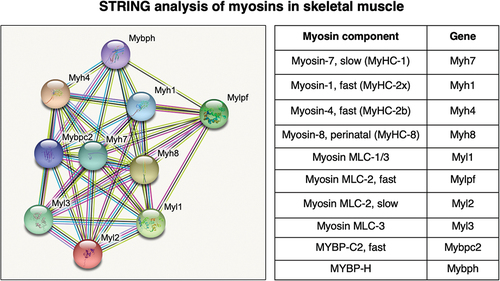
Table 1. Listing of the main cell types that are involved in embryonic myogenesis versus adult regenerative myogenesis, and their markers.
Figure 3. Proteomic profiling strategy to study skeletal muscles with a special focus on the muscle stem cell niche. Shown is the analytical workflow to study total muscle protein extracts from tissue sources, muscle-derived cell types following culturing and single-cell analysis with the help of flow cytometric sorting and separation. Frequently used methods for mass spectrometry-based protein identification and data acquisition, muscle cell analysis using stable isotope labeling and single-cell proteomics are listed. 2D-GE, two-dimensional gel electrophoresis; DDA, data-dependent acquisition; DIA, data-independent acquisition; ESI, electrospray ionization; FACS, fluorescence-activated cell sorting; FT, Fourier-transform ion cyclotron resonance; GeLC, gel electrophoresis-liquid chromatography; iBASIL, Improved Boosting to Amplify Signal with Isobaric Labeling; ICAT, isotope-coded affinity tags; ICPL, isotope-coded protein labeling; iTRAQ, isobaric tagging for relative and absolute quantitation; LC, liquid chromatography; LFQ, label-free quantification; MALDI-ToF, matrix-assisted laser desorption/ionization time-of-flight; MS, mass spectrometry; MudPIT; multi-dimensional protein identification technology; PRM, parallel reaction monitoring; PTMs, post-translational modifications; SCoPE, Single Cell ProtEomics by Mass Spectrometry; SILAC, stable isotope labeling by amino acids in cell culture; SRM/MRM, selected/multiple reaction monitoring; SWATH, Sequential Window Acquisition of all Theoretical Mass Spectra; TDA, targeted data acquisition; TMT, tandem mass tags.
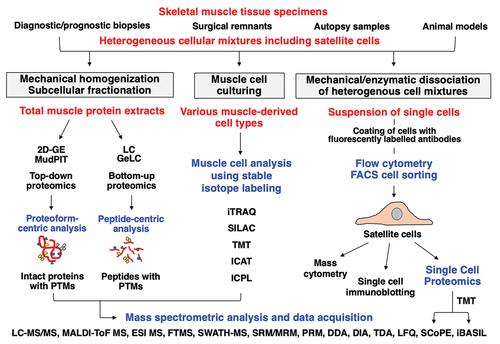
Table 2. Overview of major proteomic approaches and their bioanalytical advantages versus technical limitations.
