Figures & data
Figure 1. Heterozygous deletion of ATG5 promotes tumor growth in ApcMin/+ mice. (A) ATG5 and LC-3 protein levels in adenomas were determined by Western blotting with β-actin as a loading control. (B) Representative images of intestinal tracts from ApcMin/+ATG5+/+ and ApcMin/+ATG5+/- mice at age 6 months. (C) Micrographs of hematoxylin and eosin stained colonic tumor sections. Histological analysis showed that dysplasia, loss of nuclear polarity and complex crypt outlines with cribriform glands in adenomas of both ApcMin/+ATG5+/+ mice (a and a‘) and ApcMin/+ATG5+/− mice (b and b’). Heterozygous deletion of ATG5 had no significant effect on the malignant progression of Apc-mediated intestinal tumor. Images a‘ and b’ (×200 magnification) are high magnification of insets in a and b (×40 magnification), respectively.
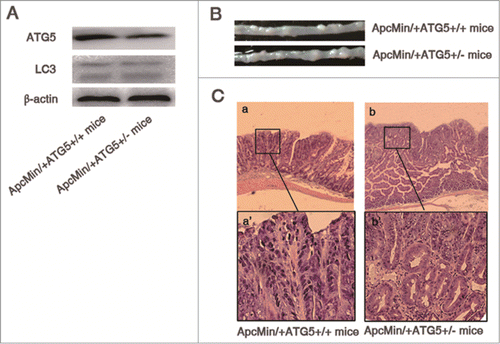
Table 1. Effect of IFN-γ on the incidence, number and size of ApcMin/+ATG5+/+ and ApcMin/+ATG5+/- intestinal adenomas
Figure 2. IFN-γ treatments effectively prevent intestinal adenomas in ApcMin/+ATG5+/− mice. (A) Representative images of intestinal tracts from ATG5 deficient ApcMin/+ mice after early treatment with IFN-γ. (B) Micrographs of hematoxylin and eosin stained colonic tumor sections. Histological analysis of intestinal adenomas in ApcMin/+ATG5+/− mice receiving vehicle revealed well-formed adenomas with severe dysplasia (a and a‘). Following early treatment with IFN-γ, adenomas of ApcMin/+ATG5+/− mice mostly exhibited hyperplastic morphology without obvious dysplasia in the polypoid area of mucosa of intestine (b and b’). Images a‘ and b’ (×200 magnification) are high magnification of insets in a and b (×40 magnification), respectively.
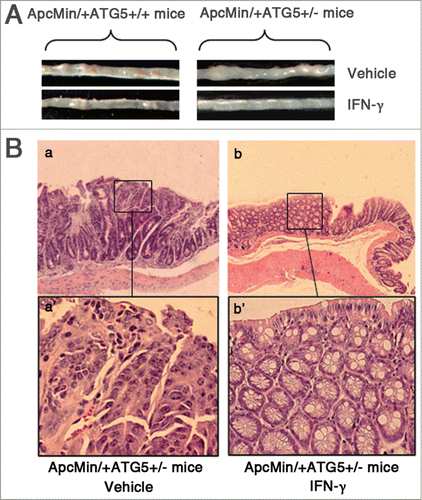
Figure 3. IFN-γ treatments do not cause any significant toxic reaction. Effects of early or late treatment of ApcMin/+ATG5+/+ mice with IFN-γ on body weight (A), average diet (B) and the number of WBCs (C) and platelets (D). *P < 0.05.
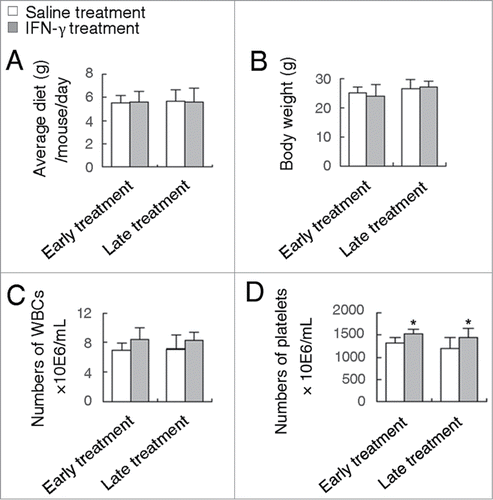
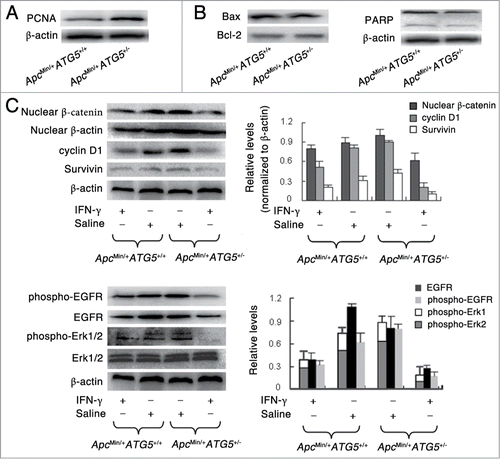
Figure 4. Heterozygous deletion of ATG5 promotes cell proliferation, activates Wnt/β-catenin and EGFR/ERK1/2 pathways and enhances the effects of IFN-γ-dependent suppression of these 2 signaling pathways. Western blotting assay showed the protein levels of PCNA (A), apoptosis-related protein including bax, bcl-2 and PARP (B), Wnt signaling-related protein including nuclear β-catenin, cyclin D1 and Survivin (C) and EGFR/Erk1/2 signaling-related protein including EGFR, phospho-EGFR and phospho-Erk1/2 (D). The protein β-actin served as a loading control and for the study involving phospho-Erk1/2, total Erk1/2 served as a control. The data presented is representative of 3 experiments. Significant differences are not shown.