Figures & data
Figure 1. CD4+CD25+Treg cells were freshly isolated from mouse splenocytes using magnetic beads. Mouse splenocytes went through negative selection for CD4+ T cells, followed by CD25+ positive selection. After the process above, the purity was up to >94% by FCM. Data was representative of at least 3 cell isolation experiments.
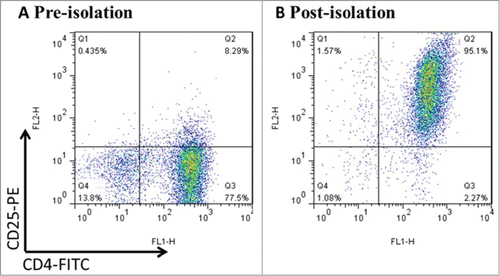
Figure 2. MENK reduced the viability of CD4+CD25+ Treg cells and inhibited TGF-β mediated conversion in vitro. (A), The viability of CD4+CD25+Treg cells was assessed by MTS. (B), Isolated CD4+CD25-T cells underwent TGF-βconversion in the presence of plate-coated anti-CD3 plus soluble anti-CD28 and IL-2 for 48 h or 72 h. The cells receiving treatment of MENK at 10−12 M or RPMI 1640 alone were examined by FCM analysis. Gate was set on CD4+CD25+, and the expression of Foxp3 was shown. (C), Real-time PCR was conducted to quantify the mRNA level of Foxp3 of the cells receiving treatment for 72 h. (D), FCM analysis for the expression pattern of CTLA-4, CCR4, GITR, and CD69 in TGF-βinduced Treg cells for 72 h. Data was investigated by Flowjo software. The red curve was the isotype control staining; the blue curve indicated specific staining. Data was collected from at least 2 independent experiments with similar results and presented as the mean±SD (triplicates). *P < 0.05; **P < 0.01 versus that in RPMI 1640 group (CONT group) as determined by Student's t test or one-way ANOVA.
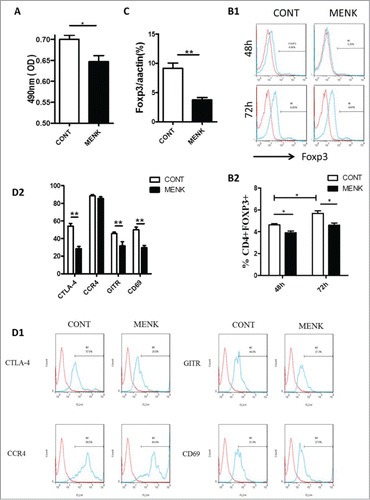
Figure 3. MENK inhibited Treg cells by affecting the phosphorylation and nuclear translocation of Smad2/3. (A),WB for analysis of phosphorylated Smad2/3. Band intensities were quantified using Quantity One version4.6.2 software. (B), The slides of cells were incubated with anti-phospho-Smad3 (1:200), and examined by Fluorescence Microscope with Digital CCD Imaging System. Analysis was done with ImageJ software. The data were representative of 3 separate experiments with similar results. The concentration for MENK treatment was 10−12 M. Data was presented as the mean±SD. *P < 0.05; **P < 0.01 vs. the RPMI 1640 group
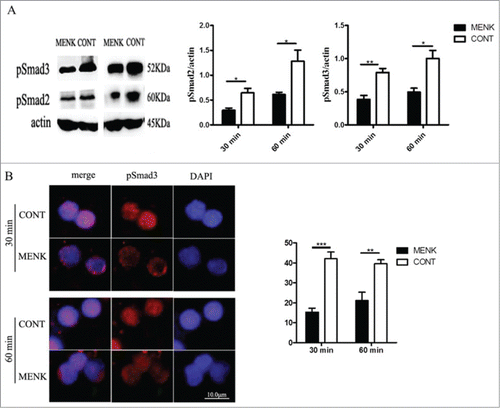
Figure 4. MENK suppressed the tumor growth. (A), C57BL/6 mice were injected with 2 × 106 S180 sarcoma cells s.c. and randomized prior to treatment (8 mice for each treatment), as indicated. MENK (20 mg/kg) was administrated i.p. daily for 14 days after tumor inoculation. Photographs of tumors were the sizes measured on day15(Original magnification × 1). (B and C), The weights and sizes of tumors. Data was presented as the mean±SD. *P < 0.05, ***P < 0.0001 versus that in control group (CONT group) as determined by Student's t test.
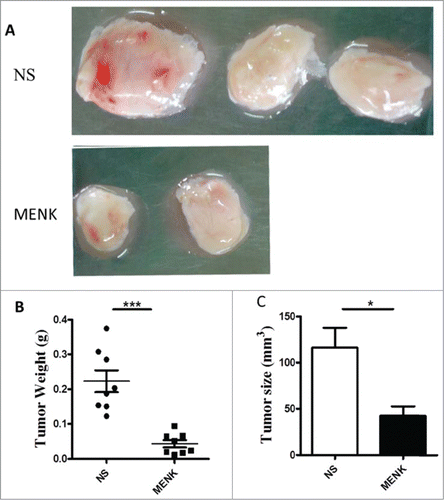
Figure 5. MENK suppressed splenic CD4+Foxp3+ Treg cells in S180 model mice. (A1 and A2), Isolated splenocytes were stained with anti-CD4 (FITC) and anti-Foxp3 (APC) and submitted to FCM analysis. (B), The mRNA level of Foxp3 in splenocytes of both groups. (C), The cells were gated on CD4+Foxp+ and analyzed for CTLA-4, GITR and FasL- positive cells, respectively. Data was analysis by FCS Express and Flowjo software. The red line was the isotype control staining; the blue line indicated specific staining mentioned above. The data were representative of at least 3 independent experiments with similar results and were presented as the mean±SD. *P < 0.05; **P < 0.01 vs. that in control group (NS group) as determined by Student's t test.
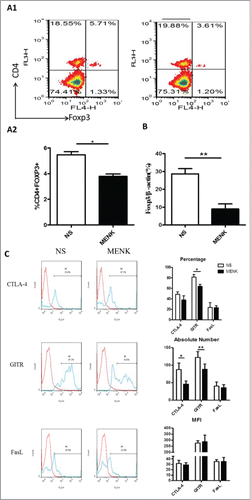
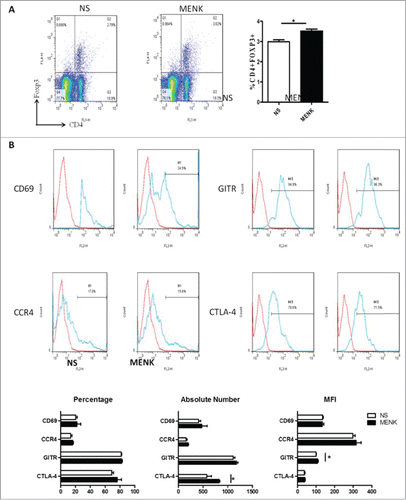
Figure 6. (A), Collected lymphocytes from tumor draining lymph nodes (TDLN) were stained with anti-CD4 (FITC) and anti-Foxp3 (APC) and submitted to FCM. (B), CD4+Foxp3+ cells were gated on and analyzed forCTLA-4, GITR, CCR4 and CD69- positive cells, respectively. The red line was the isotype control staining; the blue line indicated specific staining. The data are one representative of 3 separate experiments with similar results. Data were presented as the mean±SD. *P < 0.05 versus that in control group (CONT group) as determined by Student's t test.