Figures & data
Figure 1. (A) Crystal violet staining of ADI treated and control cells. Quantitative analysis of crystal violet stained adherent cells upon 72 h of incubation with increasing ADI concentrations. Quantification was done by measurement on a microplate reader at 570 nm (reference wavelength: 620 nm). Untreated cells were set as 100%. Results show data of 3 separate experiments. Values are given as the mean % of adherent cells ± SD. (B, C) Proliferation upon ADI exposure. CMFDA labeled cells were treated with ADI for a total of 6 days. Flow cytometric proliferation analysis was done every day of treatment. **P < 0.01 vs. control; *P < 0.05 vs. control; t-test. (B) Quantitative analysis of mean fluorescence intensity of CMFDA-labeled HROG05 cells. *ADI day 1,**ADI days 2 and 3. (C) Representative histograms showing reduced fluorescence in control cells compared to ADI-exposed cells. **P < 0.01 vs. control; *P < 0.05 vs. control; t-test.
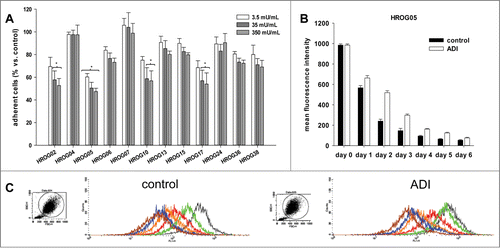
Figure 2. Expression levels and methylation profile of ASS1 and ASL in a panel of GBM cell lines. (A) Quantitative SYBR Green-1-based PCR to detect ASS1 and ASL expression in GBM cell lines. Expression was normalized to a housekeeping gene (GAPDH). Expression levels are given as 2^-dCt. (B) MSP of ASS1 and ASL as described in material and methods. Methylation profile of these 2 genes in GBM cell lines was assessed by using one primer set for ASL (Set 1) and 2 different primer sets for ASS1 (Set 2 and Set 4), representing distinct CpG islands. For ASS1, results for Set 2 are shown. (C) Quantitative MSP of CpG islands within ASS1 and ASL, respectively. Results show the methylation level of GBM cell lines with high sensitivity toward ADI. SSS1-treated leukocytes were set as 100 % and all other data were given as % vs. SSS1. Values are given as the relative expression ± SD from 3 independent replicates.
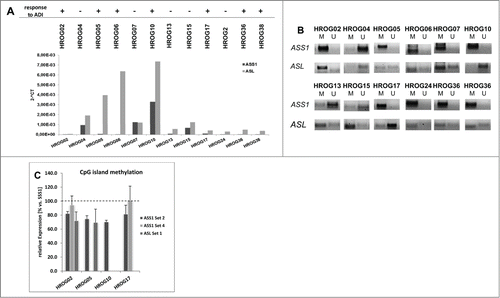
Figure 3. Crystal violet staining of GBM cell lines treated with ADI, cytostatic drugs or a combination of both. (A) Macroscopic examination of oncolytic activity. (b) Quantitative analysis of adherent cells upon 72 h and 144 h of incubation with ADI and drug (combinations). Untreated cells were set as 100% and all other data were given as % vs. control. Values are given as the mean % of adherent cells ± SD of 3 separate experiments. **P < 0.01 vs. control; *P < 0.05 vs. control; U-test.
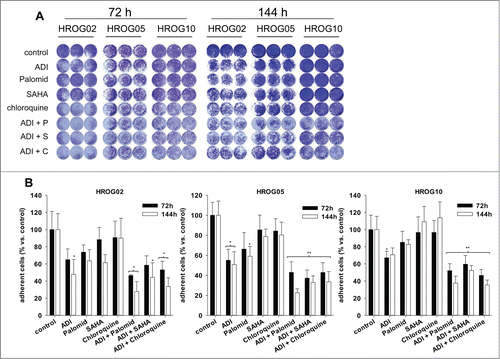
Figure 4. Cell cycle analysis of (A) HROG02 and (B) HROG05 cells. Cells were treated with ADI, selected drugs or combinations thereof for a total of 72 h. Read out was done by flow cytometry. Values are given as the mean % of cells ± SD of 3 separate experiments.
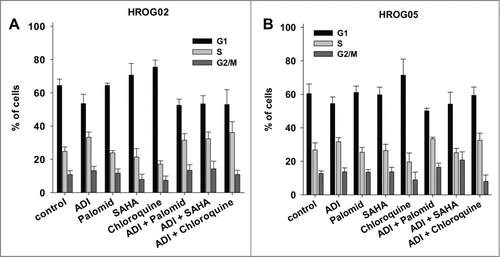
Figure 5. Sub-G1 analysis of GBM cells. Cells were treated with ADI, selected drugs or combinations thereof for 72 h and 144 h, respectively. Read out was done by flow cytometry using single gating strategy. All cells in Gate 1 were measured (20,000 events in total). (A) Representative dot plots of HROG05 treated cells. (B) Quantitative analysis of cells with Sub-G1 Peak. Values are given as the mean% of cells ± SD of 3 separate experiments.
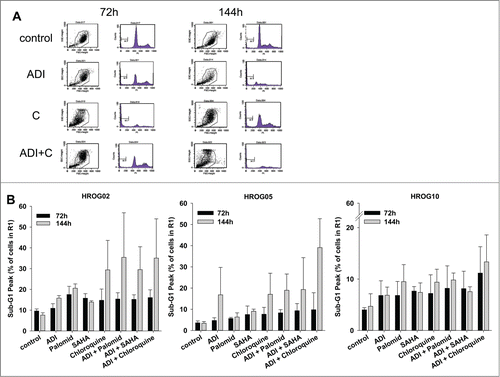
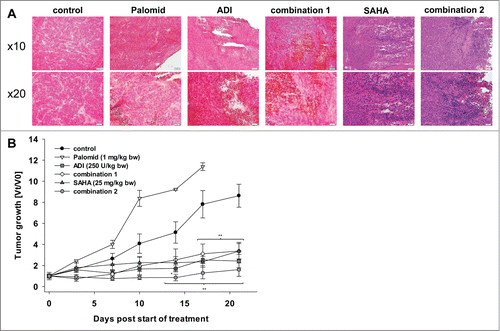
Figure 6. HROG05 histology and xenograft growth. Therapy was performed by repetitive local application of ADI (250 U/kg bw), Palomid (1 mg/kg bw), SAHA (25 mg/kg bw) and a combination of either Palomid 529 + ADI (=combination 1) or SAHA + ADI (=combination 2). Each therapy was given twice a week (a total of 6 injections) (n=5 mice per group). Tumor-carrying control animals received equivalent volumes of PBS/DMSO (n = 5 per group). (A) Representative HE staining of HROG05 xenografts. Pictures of 4 μm slides were taken at low and high resolution, respectively. (B) Tumor growth curve. Tumor volumes are given as x-fold increase vs. day 0 (Vt/V0) (start of treatment) ± SD. *P < 0.05 vs. control; **P < 0.01 vs. control.