Figures & data
Table 1. Primer sequences for qRT-PCR amplification of different genes
Table 2. Comparison of FOXQ1 expression in tumor and normal tissues
Table 3. Correlation between FOXQ1 expression and clinical parameters of colorectal cancer patients
Figure 1. FOXQ1 was overexpressed in colorectal tumor cells. FOXQ1 was stained in colorectal cancer samples (A), adjacent nontumorous tissues (B), and distal normal colorectal mucosa samples (C). Images were acquired at 200X. Representative images are shown.
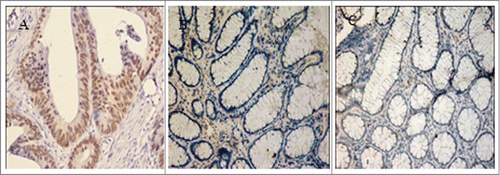
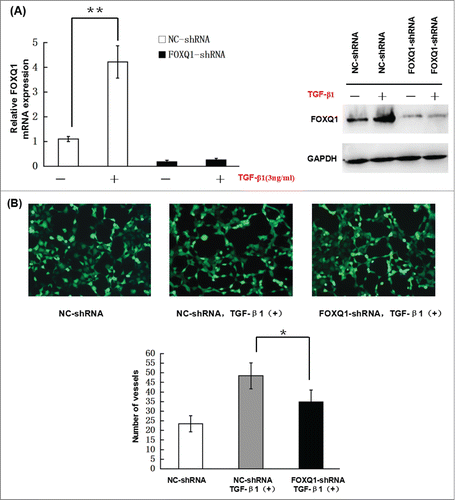
Figure 2. Knockdown of FOXQ1 expression by shRNA. (A) SW480 cells were transduced with lentivirus containing FOXQ1-sh1, -sh2, -sh3, or NC-shRNA for 72 hours. GFP signals were detected by fluorescence microscopy (100X) to indicate the transduction efficiency. (B) FOXQ1 mRNA (left panel) and protein (right panel) levels in SW480 cells transduced with FOXQ1-sh1, -sh2, -sh3, and NC-shRNA were measured by qRT-PCR and Western blot, respectively. (C) Knockdown efficiency of FOXQ1-sh1 was confirmed by qRT-PCR (left panel) and Western blot (right panel). The NC-shRNA transduced cells (NC-shRNA) and non-transduced cells (control group) were used as controls. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.
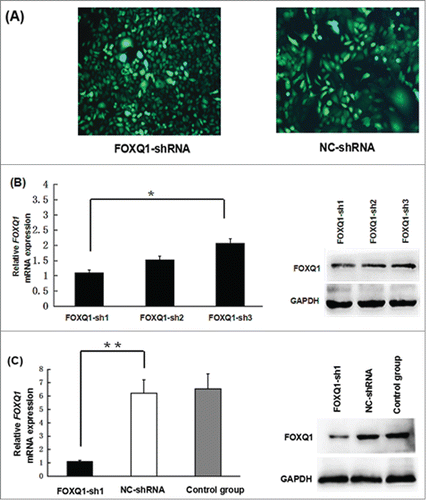
Figure 3. Knockdown of FOXQ1 reduced the aggressive tumor behaviors of SW480 cells. (A) An in vitro angiogenesis assay measured the vessel formation of NC-shRNA-transduced EA.hy926 cells growing in FOXQ1- and NC-shRNA cell culture media after 48 hours. The number of vessels was counted by fluorescent microscopy (120X). (B) FOXQ1- and NC-shRNA cells seeded in the upper chamber of Transwell were examined by optical microscopy (120X) (upper panel). After 36 hours, the cells migrating through the insert membrane between the upper and lower chambers were fixed and stained by crystal violet. The number of migrated cells was counted by optical microscopy (120X) (lower panel). (C) FOXQ1- and NC-shRNA cells were treated with 5-FU (left panel) and L-OHP (right panel) at 100 μg/ml for 24 hours. Annexin and 7-AAD were used in flow cytometry to measure the percentage of apoptotic cells. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.
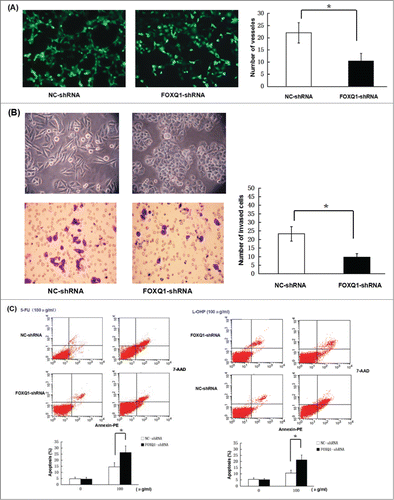
Figure 4. Knockdown of FOXQ1 expression suppressed Wnt signaling. (A) NC- and FOXQ1-shRNA cells were stained with fluorescent β-catenin antibody (red) and DAPI nuclear stain (blue). (B) Western blot analysis of the protein level of β-catenin in NC- and FOXQ1-shRNA cells. (C) Left panel, qRT-PCR analyses of the mRNA levels of Wnt downstream target genes and EMT associated markers in NC- and FOXQ1-shRNA cells; right panel, Western blot analyses of the protein levels of Wnt downstream target genes and EMT-associated markers in NC- and FOXQ1-shRNA cells. The results are expressed as mean ± SD **P < 0.01.
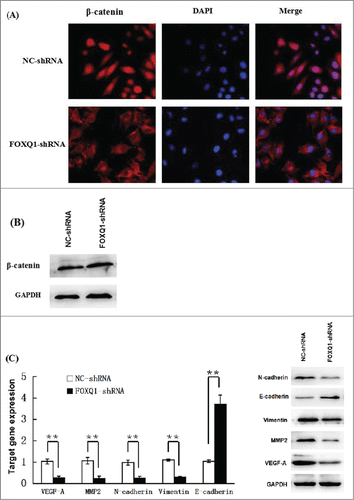
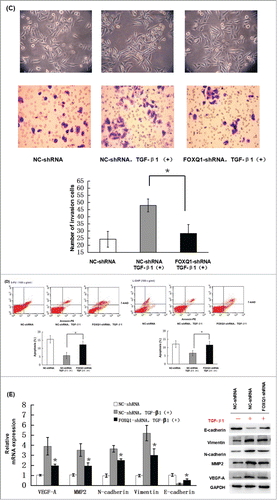
Figure 5. Knockdown of FOXQ1 partially inhibited TGF-β1-induced aggressive tumor behaviors in SW480 cells and the changes in expression of downstream genes. (A) NC- and FOXQ1-shRNA cells were incubated in growth media supplemented with TGF-β1 (3 ng/ml) for 3 days. FOXQ1 expression was measured by qRT-PCR and Western blot. (B) The culture supernatant from NC-shRNA cells cultured for 3 days or NC- and FOXQ1-shRNA cells cultured for 5 days containing TGF-β1 (3 ng/ml) was collected and used in an in vitro angiogenesis assay. The vessel formation was measured in NC-shRNA-transduced EA.hy926 cells. The number of vessels was counted by fluorescent microscopy (120X). (C) FOXQ1- and NC-shRNA cells were treated with TGF-β1 (3 ng/ml) for 5 days, and cell morphology was examined by optical microscopy (120X) (upper panel). For the invasion assay, FOXQ1- and NC-shRNA cells were pre-treated with TGF-β1 (3 ng/ml) for 3 days before being seeded in the upper chamber of Transwell. After 36 hours, the cells migrating through the insert membrane were fixed and stained by crystal violet. The number of migrated cells was counted by optical microscopy (120X) (lower panels). (D) For the drug sensitivity assay, FOXQ1- and NC-shRNA cells were incubated in TGF-β1-containing media (3 ng/ml) for 4 days, and 5-FU or L-OHP (100 μg/ml) was added to the cell culture for an additional 24 hours. Flow cytometry was performed to measure cell apoptosis. (E) Expression of Wnt downstream target genes and EMT-associated markers in TGF-β1 treated NC- and FOXQ1-shRNA cells were measured by qRT-PCR and Western blot. The results are expressed as mean ± SD *P < 0.05, **P < 0.01.