Figures & data
Table 1. Association of UHRF1 expression in the tumor tissues with demographic and clinicopathologic characteristics in 238 patients with GC
Table 2. Univariate Analysis and Multivariate analysis of the Correlation Between Clinicopathological Parameters and Survival of Patients With Gastric Cancer
Table 3. Proliferation related genes upregulated after UHRF1 downregulation in MKN45 cells.
Figure 1. Kaplan–Meier survival curves of GC patients with different level of UHRF1 expression stratified by the TNM stage of the tumor (log-rank test). (A) Correlation of UHRF1 expression with overall survival (cum survival) in all stages. (B) Correlation of UHRF1 expression with overall survival in Stage I–II. (C) Correlation of UHRF1 expression with overall survival in Stage III–IV.
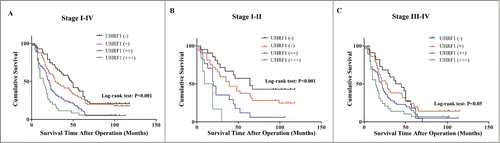
Figure 2. Expression of UHRF1 in GC cell lines. (A) The expression level of UHRF1 mRNA in 3 GC cell lines and one immortalized normal gastric mucosal epithelial cell line (GES) was measured using qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (B) The expression of UHRF1 in 3 GC cell lines and GES cell line was examined through western blot analysis. β-actin was used as an internal control.
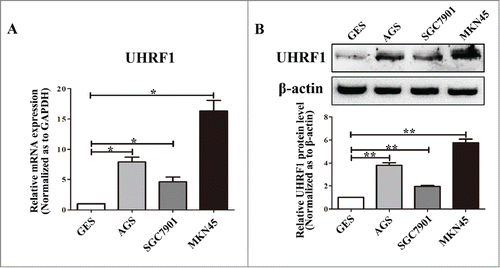
Figure 3. UHRF1 promotes GC cell proliferation in vitro. (A) Western blot analysis of UHRF1 expression in MKN45 cells infected with UHRF1 shRNA (shUHRF1) or negative control (shNC) and parental cells. β-actin was used as an internal control. (B) Western blot analysis of UHRF1 in SGC7901 cells transfected with UHRF1 plasmid or vector control (NC) and parental cells. (C) XTT assay of MKN45, MKN45-shNC and MKN45-shUHRF1 cells. (D) XTT assay of SGC7901, SGC7901-NC and SGC7901-UHRF1 cells. (E) Colony formation assay of MKN45 cells infected with shUHRF1 and SGC7901 cells transfected with UHRF1 plasmid. Colonies were evaluated and values were reported as the ratio. (F) Apoptosis assay of MKN45 cells infected with shUHRF1 and SGC7901 cells transfected with UHRF1 plasmid. (G) Flow cytometry cell cycle analysis of MKN45, MKN45-shNC and MKN45-shUHRF1 cells. (H) Flow cytometry cell cycle analysis of SGC7901, SGC7901-NC and SGC7901-UHRF1 cells. Data are shown as mean ± SEM (n = 3) of one representative experiment. Similar results were obtained in 3 independent experiments.
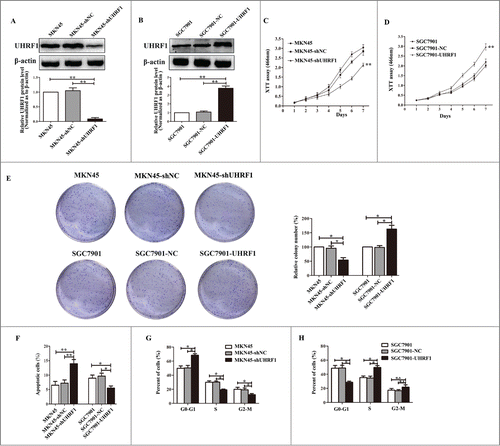
Figure 4. UHRF1 reduces tumor suppressor expression via enhancing methylation. (A) Effects of UHRF1 silencing on gene methylation of 7 tumor suppressor genes in MKN45 cells infected with UHRF1 shRNA (shUHRF1) assayed by Methylation-specific PCR (MSP). (B) Effects of UHRF1 upregulation on gene methylation of 7 tumor suppressor genes in SGC7901 cells transfected with UHRF1 plasmid assayed by MSP. (C) Effects of UHRF1 silencing on mRNA expression of 7 tumor suppressor genes in MKN45 cells infected with shUHRF1 assayed by qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (D) Effects of UHRF1 upregulation on mRNA expression of 7 tumor suppressor genes in SGC7901 cells transfected with UHRF1 plasmid assayed by qRT-PCR.
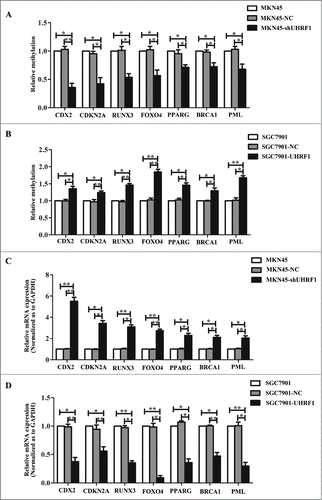
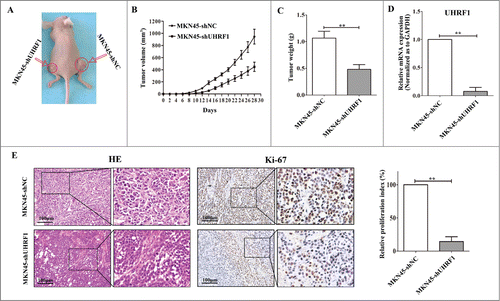
Figure 5. Down-regulation of UHRF1 inhibits GC growth in vivo. (A) MKN45-shUHRF1 and MKN45-shNC cells were injected subcutaneously into nude mice. At 4 weeks after implantation, MKN45-shUHRF1 cells produced smaller tumors than control cells. (B) Growth curve of tumor volumes. Each data point represents the mean ±SEM of 5 mice. (C) Average weight of tumors in nude mice. (D) The mRNA expression of UHRF1 in transplanted tumors formed by MKN45-shUHRF1 and MKN45-shNC cells was measured using qRT-PCR. GAPDH was used as an internal control and the fold change was calculated by 2−ΔΔCt. (E) Representative photographs of H&E staining and immunohistochemical analysis of Ki-67 antigen in tumors of nude mice (left). Comparison of proliferation index is shown on the right.