Figures & data
Figure 1. Results of toxicity experiment of MEB55 (12.5, 25, 50, 100 and 150 mg/kg) in Hsd:ICR(CD-1®) mice (n = 5 mice/treatment). Additional group was set as vehicle control. Mice were injected intraperitoneal (IP) twice a week with the MEB-55 solutions, dissolved with DMSO:Chromophor®EL 1:1 and diluted with double distilled water to the required concentration. (A) Body weight (% of time 0) at 8 and 14 d post injection (DPI). Values are means ± SE. (B) Hematoxylin and eosin (H&E) staining of liver sections. * - tissue abnormalities; pictures taken at different magnifications are shown (×100, ×400). (C) Apoptosis cell staining of liver sections using DeadEndTM Fluorometric Tunnel system kit. White arrows point to the apoptic cells. (D) Immunostaining for macrophages of liver sections using polyclonal anti-macrophage antibodies (CL194P). Yellow arrows mark area of increase in macrophage presence.
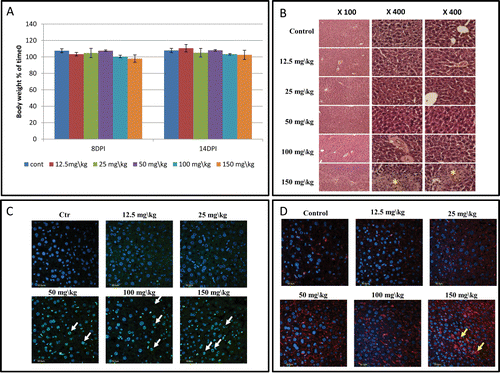
Figure 2. The effect of MEB55 (25 mg/kg) or vehicle treatments on xenografts of MDA-MB-231 in animal model of nude BALB/cOlaHsd-Foxn1nu mice. (A) Tumor volume (calculated as V = π(length)×(width)×(height)/6) in mice treated with MEB55 (25 mg/kg) or vehicle; significant differences were analyzed by Wald test. * - Means are significantly different as determined by Student's-t-test (P < 0.05). (B) Tumor growth rate (0.5 × small diameter2 × large diameter/day) in mice treated with MEB55 (25 mg/kg) or vehicle over DPI (days post injection). Values are means ± SE (n = 8); Different letters above the bars indicate statistically significant differences between means determined by Student's-t-test (P ≤ 0.05).
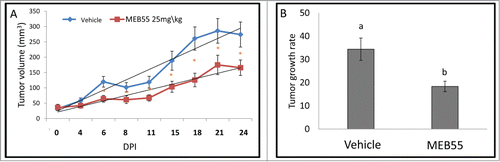
Figure 3. Dose response curves of MDA-MB-231 cell viability following treatment with MEB55 alone and in combination with paclitaxel. Cells were exposed to either single agent drug (A) paclitaxel or (B) MEB55 (circle) or to drugs combinations (A) paclitaxel + 7.5 µM of MEB55 or (B) Meb55 + 10 nM of paclitaxel (square) for 48 hours followed by XTT assay to determine cell viability. Cell survival (%) of control is calculated by = 100 × (At-Ac)(treatment) / (At-Ac)(control), where At and Ac are the absorbencies (450nm) of the XTT colorimetric reaction in treated and control cultures respectively minus non-specific absorption measured at 650nm. Data points are connected by non-linear regression lines of the sigmoidal dose-response relation. Values are means ± SE of 3 independent experiments. IC50 values were determined with nonlinear regression analysis. * Means are significantly different between single agent and combination as determined by Student's-t-test (P ≤ 0.05).
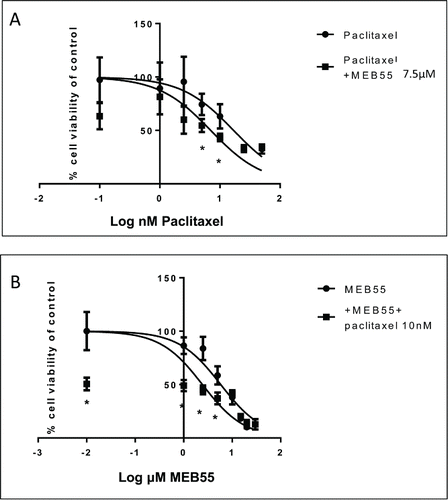
Figure 4. The effect of MEB55 (25 mg/kg; A) or paclitaxel in 2 concentrations (15 or 7.5 mg/kg) or combination of MEB (25 mg/kg) and paclitaxel (7.5 mg/kg) treatments on xenografts of MDA-MB-231 in animal model of nude BALB/cOlaHsd-Foxn1nu mice. Administration was by IP twice a week for 3 weeks. Tumor volume (mm3; calculated as V = π(length)×(width)×(height)/6) at the end of experiment (12 d post treatment initiation) is shown. Values are means ± SE (n = 8). *, Means are significantly different or NS, means are not-significantly different, as determined by Student's-t-test (P ≤ 0 .05). £, Treatment significantly different compared to control.
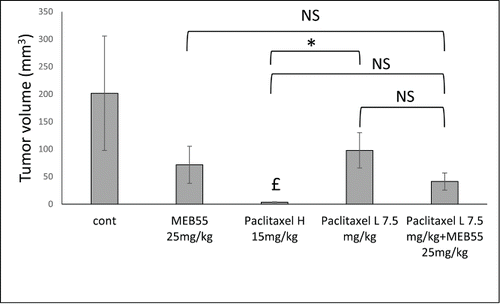
Figure 5. Fluorescent images of MDA-MB-231 cancer cells immunostained for α-tubulin (Green staining- Alexa Fluor 488) following MEB (15 µM), ST362 (15 µM), paclitaxel (Taxol, 2.5 nM), vehicle control treatments or untreated control for 5 hr. Blue nuclei staining- DAPI. Images were taken using Zeiss Axiovert 200M Fluorescense inverted microscope at ×630 magnification.
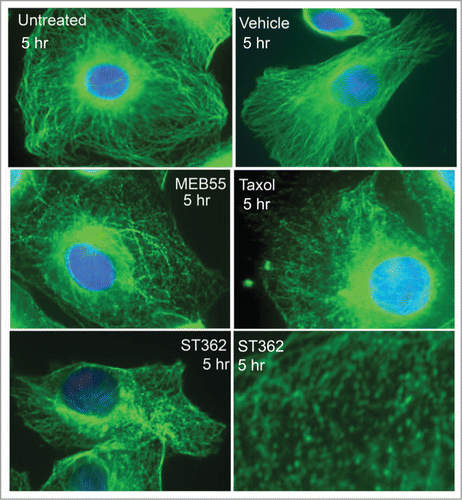
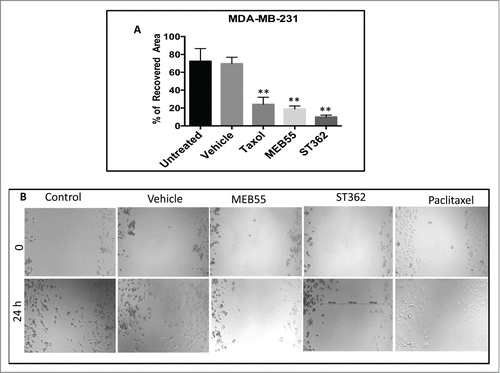
Figure 6. The effect of MEB55 (7.5 µM), ST362 (7.5 µM) or paclitaxel (Taxol, 2.5 nM), vehicle and untreated control on recovered area of confluent monolayers of MDA-MB-231 cancer cell line. Monolayer was scraped in a straight line and % recovered area of the scratch by cells that migrated into the scratch was measured from images taken at time 0 and 24 h and calculated as (0 h cell free area in treatment - 24 h cell free area in treatment)/ (0 h cell free area in untreated - 24 h cell free area in untreated) × 100. (A) Values are means ± SE (n = 8). *, Means are significantly different as determined by Student's-t-test. Paclitaxel and MEB55 treatments (P < 0.0019); ST362 treatment (P < 0.0004). (B) An example for the effect of MEB55 (7.5 µM), ST362 (7.5 µM) and paclitaxel (2.5 nM), vehicle and untreated control on recovered area of confluent monolayers of MDA-MB-231 cancer cell line cells in comparison to vehicle and untreated control. Bars demonstrate length of measurements.