Figures & data
Figure 1. DSB repair pathways are hyperactivated in Hep3B and HuH7 cells. (A) Reporter constructs for analysis of DSB repair. The HR reporter comprises 2 inactivated copies of GFP-Pem1. In the first one, a 22 nt and an insertion of 2 I-SceI recognition sites in inverted orientation are inserted into the first exon of GFP. In the second copy, both the ATG and second exon of GFP are deleted. Upon induction of DSBs by I-SceI, only gene conversion restores GFP expression. The starting NHEJ reporter is also GFP negative as it contains a GFP gene with an intron from the rat Pem1 gene and an adenoviral exon (Ad). Two I-SceI recognition sites flank the Ad exon in an inverted orientation. Upon induction of a DSB, only NHEJ can turn cells GFP+. SA, splice acceptor; SD, splice donor. (B) Analysis of HR or NHEJ efficiency in 4 liver cell lines. At 48 h post splitting, cells were co-electroporated with I-SceI linearized 0.6 μg HR or 0.2 μg NHEJ reporter plasmids and 0.005 μg of the DsRed expression vectors for normalizing transfection efficiency. Three days later, cells were harvested for FACS analysis. The ratio of GFP+ versus DsRed+ cells was used as a measure of repair efficiency. All the experiments were repeated more than 3 times and all results were normalized with that of Chang liver. Error bars stand for standard deviation (SD).
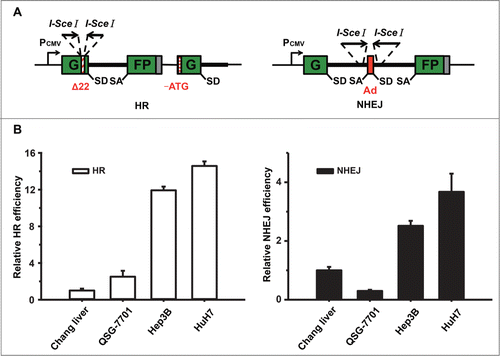
Figure 2. Harmine inhibits the efficiency of HR and suppresses the proliferation in Hep3B and HuH7 cells. (A) Chemical structure of Harmine. (B) Effect of Harmine on HR or NHEJ efficiency. Hep3B and HuH7 cells were pretreated with indicated concentrations of Harmine for 6 h, followed by co-transfection with linearized HR or NHEJ reporter constructs and DsRed expression plasmid. Twenty-four h later, cells were analyzed by flow cytometry. All results were normalized with that of control. (C) Effects of Harmine on the proliferation of Hep3B and HuH7. After treated with Harmine for 12 h, 24 h and 48 h, the numbers of cells were determined by MTT assay. The IC50 value at each time point was calculated using Dose-Effect Analysis software. (D) Cytotoxicity of Harmine on Chang liver or QSG-7701 cells lines. The procedure was the same as described in Figure 2C. (E and F) The combinatory treatment of Harmine and Nu7441. Hep3B or QSG-7701 cells were plated in 96-well plate and exposed to serial concentrations treatments of Nu7441 and Harmine either alone or in combination for 24 h. Cell viability was then determined by MTT assay. All experiments were repeated at least 3 times. Error bars represent SD
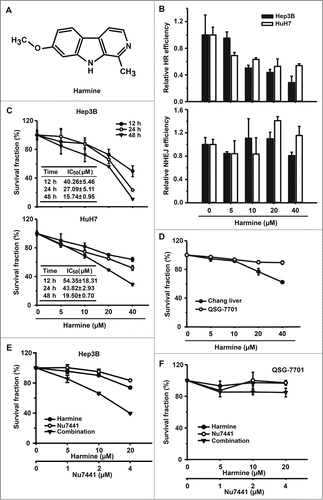
Figure 3. Harmine blocks HR by inhibiting Rad51 recruitment. (A and B) Harmine does not affect the expression levels of HR related proteins. Hep3B cells were treated with indicated concentrations of Harmine, followed by x-ray irradiation. The expression levels of HR related proteins were analyzed by Western blot. (C) Harmine disrupts the formation of Rad51 foci after DNA damage. Hep3B cells pretreated with 40 μM Harmine for 2 h were irradiated with 8 Gy X-ray. At 6 h post irradiation, Rad51 foci formation was examined by immunofluorescence. The percentage of cells containing Rad51 foci (>5 ) was displayed. **, p < 0 .01. All experiments were repeated 3 times and at least 50 nuclei were examined and quantified for each time. (D) Harmine upregulates γ-H2Ax level in Hep3B cells. Cells were treated with Harmine at various concentrations for 6 hours before whole cells lysates were extracted and analyzed by Western blot. (E) The p-Chk1 level is upregulated in Hep3B cells following Harmine treatment. The experimental procedure is as described in . (F) Harmine treatment leads to a persistent DNA damage signaling after x-ray irradiation. Hep3B cells were exposed to X-ray at 8 Gy, followed by treatment with 40 μM Harmine for 2 h. Then γ-H2Ax foci were immune-stained. The percentage of cells positive for γ-H2Ax foci (>10 ) was shown. **, p < 0 .01. All experiments were repeated 3 times and at least 50 nuclei were examined and quantified for each time.
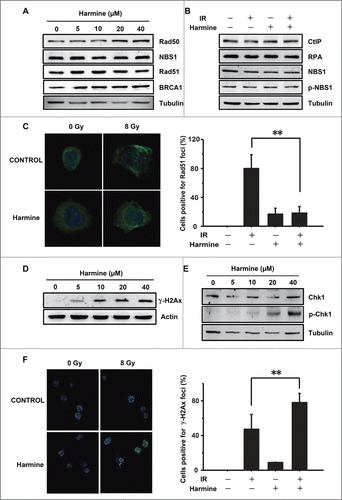
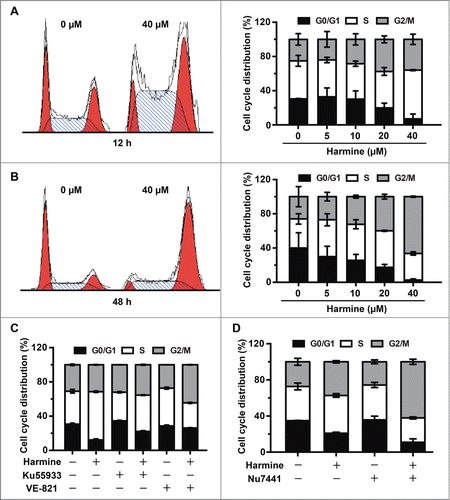
Figure 4. Harmine induces S and G2/M phase arrest in Hep3B cells in an ATM and ATR dependent manner. (A and B) Harmine arrests cells in S and G2/M phases. Hep3B cells were grown in the absence or presence of Harmine for 12 h or 48 h, respectively. Cell cycle distributions were examined by flow cytometry after PI staining. Results were representative of 3 independent experiments. (C) ATM or ATR inhibition abrogates the Harmine induced cell cycle arrest. Hep3B cells pretreated with 10 μM Ku55933 or VE-821 for 2 h were further treated with 40 μM Harmine for another 12 h. Cell cycle profiles of Hep3B cells were then detected by PtdIns staining and flow cytometry. (D) The combinatory treatment of Harmine and Nu7441 promotes cell cycle arrest of Hep3B cells. Cell cycle distributions of Hep3B cells after treatment with 4 μM Nu7441 and 20 μM Harmine either alone or in combination for 24 h were determined by PI staining followed by flow cytometry.