Figures & data
Figure 1. BRAFT1799A. status of cell lines. Allelic discrimination plot for BRAFT1799A SNP. All samples were auto-called for BRAF status (95% Confidence Interval). X-axis: wild type allele; Y-axis: mutant allele.
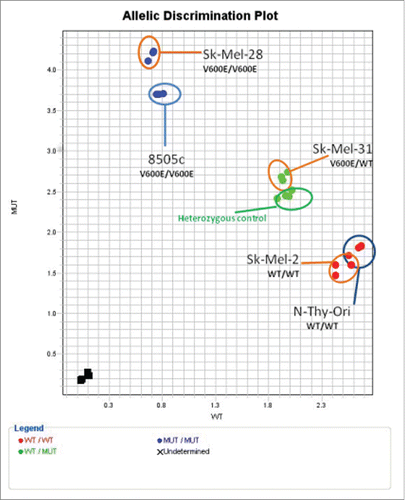
Table 1 Optimised Experimental protocols for PD0325901 MEKi Treatment.
Figure 2. MEKi resistance induces differential gene expression of key members of the MAPK Pathway in BRAFV600E cancer. Cells treated with PD0325901 (resistant) or media-alone (naïve) were analyzed for gene expression. Fold change (RQ) expression in resistant samples was normalized to RQ of paired naïve samples and averaged across biological replicates. Statistical significance (*). Error bars: SEM. X-axis: Cell Line; Y-axis: Log2 (Avg RQ Resistant/RQ Naïve) Confocal analysis for tERK1/2 detects no change between Naïve and Resistant 8505C or NThy-Ori samples. pERK1/2 can be detected in all 3 layers of the cell in naïve samples whereas it is only detected in the apical and midsection in MEKi resistant 8505C samples. Conversely in NThy-Ori: pERK1/2 is only detected in the apical and midsection in naïve samples whereas pERK1/2 can be detected in all 3 layers in MEKi resistant samples. Red staining pERK1/2, green staining total ERK1/2, blue staining nuclear (DAPI).
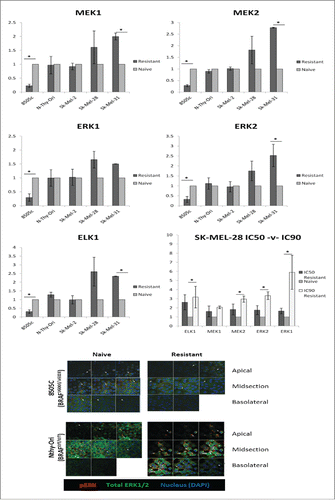
Table 2 Significant MIRNAs associated with samples.
Table 3 Association between top 20/cluster most significant GO terms.
Figure 3. Holoclones have an upregulation of stemness factors. SK-MEL-28 and 8505C holoclones were compared to their parental cell lines for expression of pluripotency and EMT genes. RQ expression in holoclone samples was normalized to RQ of parental samples and averaged across biological replicates. Statistical significance (*). Error bars: SEM. X-axis: Assayed genes; Y-axis: Log2(Avg RQ Resistant/RQ Naïve).
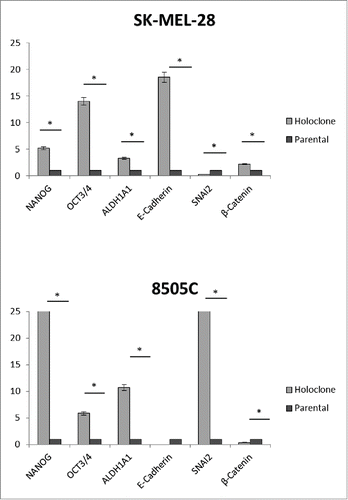
Table 4 MIR302/373/374/520 embryonic stem cell specific cell cycle regulating (ESCC) microRNA family in holoclones compared to parental cells (relative fold change +/−2).
Figure 4. MEKi resistance induces upregulation of stemness-associated genes in BRAFV600E melanoma cell lines. Cells treated with PD0325901 (resistant) or media-alone (naïve) were analyzed for gene expression. RQ expression in resistant samples was normalized to RQ of paired naïve samples and averaged across biological replicates. Statistical significance (*). Error bars: SEM. X-axis: Assayed cell line; Y-axis: Log2(Avg RQ Resistant/RQ Naïve).
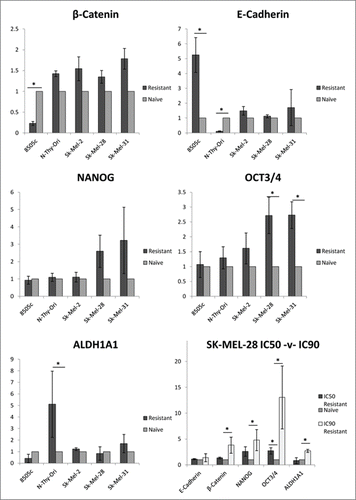
Figure 5. Protein expression is differentially regulated in SK-MEL-28 and SK-MEL-2 MEKi resistant protein samples. (A-D) Western blot and autoradiograpgy intensity analysis was performed for PD0325901 resistant and naïve samples. X-axis: Cell Line; Y-axis: Intensity target protein/Intensity Actin.
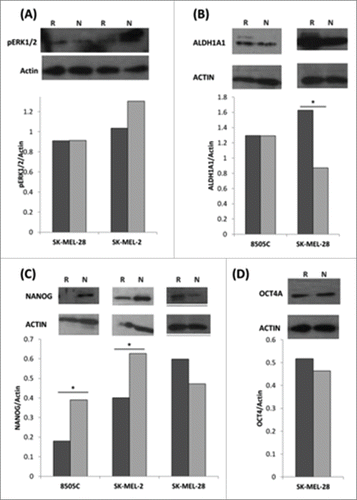
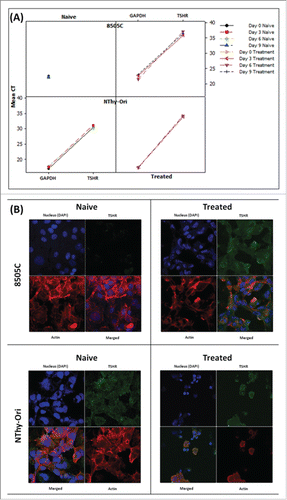
Figure 6. MEKi treatment induces reinstatement of TSHR protein in anaplastic thyroid cells (A) Gene expression analysis can detect TSHR expression in 8505C cells only following sustained MEK inhibitor treatment. TSHR is readily detectable in both the naïve and resistant BRAFWT NThy-Ori cell line. (B) Confocal analysis detects expression of the TSHR protein expression in 8505C cells following sustained MEK inhibitor treatment. TSHR protein is readily detectable in both the naïve and resistant BRAFWT NThy-Ori cell line. Confocal Legend: TSHR: Green; Actin: Red; Nucleus: Blue Images (individual color channels and merged) taken on 63X objective lens.