Figures & data
Figure 1. P1446A-05 strongly reduces proliferation of human melanoma cell lines in 2D and 3D culture. (A) P1446A-05 exhibits antiproliferative effects across different melanoma genotypes and phenotypes. Sensitivity ranking placed cutaneous melanoma lines ahead of uveal melanoma lines. Cells were incubated with a range of P1446A-05 concentrations for 72 hours, and cell viability was analyzed using the CellTiter-Glo luminescence assay. GI50 values were calculated using nonlinear regression curve fit in GraphPad Prism 6 (error bars show 95% CI). (B) Escalating doses of P1446A-05 demonstrate strong inhibition of 3D melanoma spheroids. Mel Juso cells were cultured as spheroids in matrigel and micrographs were taken after 72 hours of drug treatment. Representative images are shown; scale bars represent 50 µm in spheroid images. Analysis of spheroid size was used to calculate % inhibition relative to control.
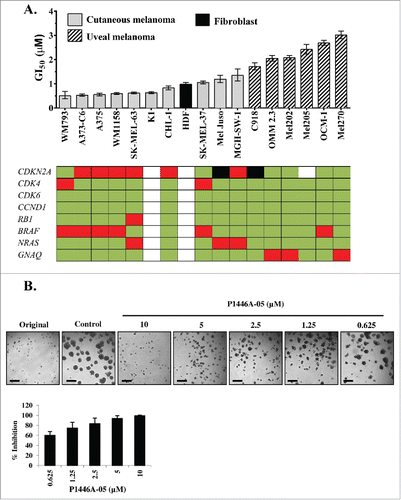
Figure 2. P1446A-05 induces cell cycle arrest and apoptosis. (A, B) P1446A-05 causes G2-phase cell cycle arrest and an increase in sub-G1-phase cells at 24 and 48 hours. Cells were treated with 1 µM and 5 µM P1446A-05 for 24 hours and 1 µM P1446A-05 for 48 hours. Cell cycle progression was assessed using PI staining and flow cytometry. (C) P1446A-05 induces apoptosis, which correlates to the increase in sub-G1-phase cells. A375 and SK-MEL-63 cells were treated with 1 µM P1446A-05, and OCM-1 cells were treated with 1.5 µM P1446A-05. Apoptosis was assessed using annexin VPI staining and flow cytometry. (D) Compared to DMSO-treated controls, A375 cells treated with 1 µM P1446A-05 revealed a robust increase in cleaved PARP. GAPDH served as a loading control. Error bars represent SEM from triplicates. *P < 0.0001, #P < 0.001.
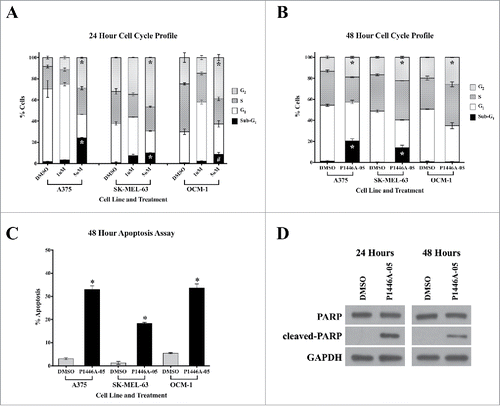
Figure 3. Intracelluar target validation. P1446A-05-mediated inhibition of CDK4 and CDK9 was confirmed via protein gel blotting for relevant phosphotargets in both CDK pathways. CDK4s phosphotarget on RB protein (ser780) becomes inhibited by 24 hours across all 3 cell lines in a dose dependent manner. CDK9s phosphotarget on RNAPII's Rpb1 (ser2) revealed better inhibition at 6 hours, and was only inhibited with the highest dose of P1446A-05 at 24 hours. The housekeeping protein GAPDH served as a loading control.
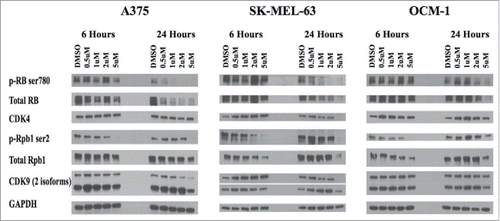
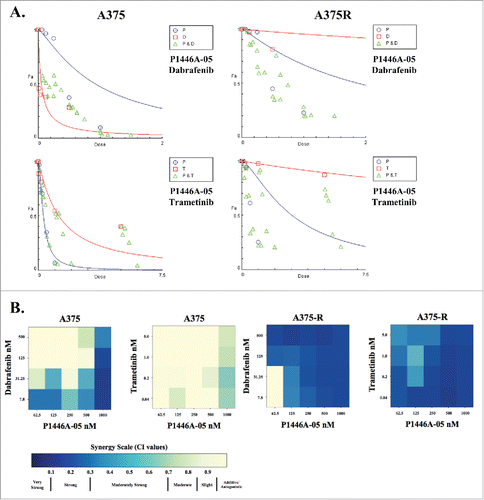
Figure 4. Synergism between P1446A-05 and other small molecule inhibitors. (A) Isobolograms demonstrating synergy between P1446A-05 and dabrafenib or trametinib. The isobolograms were generated using Compusyn software. P, P1446A-05; T, trametinib; D, dabrafenib. (B) Heat maps depicting combination indices (CI's) for dose combinations of P1446A-05 with dabrafenib and trametinib. A375 and K4 cell lines, both sensitive and resistant (A375-R, K4-R) to vemurafenib, were treated. Cells were incubated with all drug combinations at indicated doses for 72 hours, cell viability was analyzed with CellTiter-Glo, and CI's were calculated using CompuSyn software.