Figures & data
Figure 1. Screening of FDA-approved drugs for TRAIL-sensitizers in GBM cells (A) Scatter plot of U87MG cell viability upon 24 hours of treatment with drug library composed of 1200 compounds. Each dot represents the percent cell viability compared to untreated control samples. Horizontal dashed lines depict the range of viability within 3 SD of the mean viability (64 % and 112%). Red dots are drugs that have significant effect on cell viability as single agents. (B) Scatter plot of U87MG cell viability upon 24 hours of treatment with drug library composed of 1200 compounds and TRAIL together. Each dot represents the combinatorial effect of the individual drug (5 µM) and TRAIL (25 ng/ml) on cell viability compared to untreated control samples. Horizontal dashed lines depict the range of viability within 3 SD of the mean viability (29% and 83%). Red and blue dots indicate the combination effect below the threshold of 3 SD from the mean cell viability. (C) Top 26 hits were determined via cut off value of 29% cell viability after drug and TRAIL addition. DMSO and Bortezomib, negative and positive control respectively, are indicated in gray bars. The effect of drug and TRAIL are grouped among hits, as drugs with significant effect on cell viability as single agents (red bars) and the others (blue bars). Error bars represent “mean ± NSEM.” SD: standard deviation from mean cell viability.
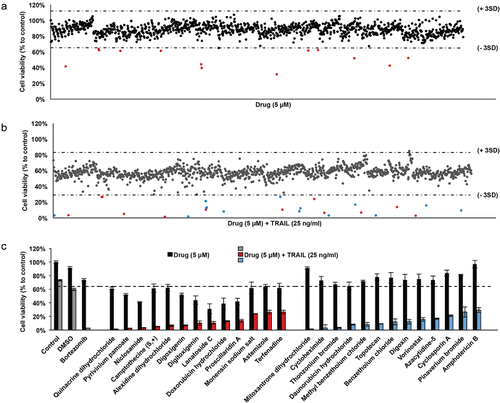
Table 1. Top hits of U87MG and U87MG-R50 drug screens. Drugs that have significant effect on GBM cells as single agents are indicated in bold.
Figure 2. Effect of a cardiac glycoside, Digitoxigenin, on GBM and non-malignant cell viability. Cell viability detection after 24 hours of Digitoxigenin and/or TRAIL addition. (A) The effect of different doses of Digitoxigenin and TRAIL combinations on U87MG. (B) The chosen dose combination of 500 nM Digitoxigenin and 50 ng/ml TRAIL on BJ and NHA. (C) Low doses of Digitoxigenin (1–50 nM) tested on a panel of human fibroblasts: KUFibro4, KUFibro5, KUFibroMNG, BJ and Human Astrocytes (NHA). (D) Doses up to 10 nM were combined with 50 and 100 ng/ml TRAIL on BJ cells. (E) Low dose combination of Digitoxigenin on U87MG cells. * indicates p <0.05 and ns indicates nonsignificant (p>0.05).
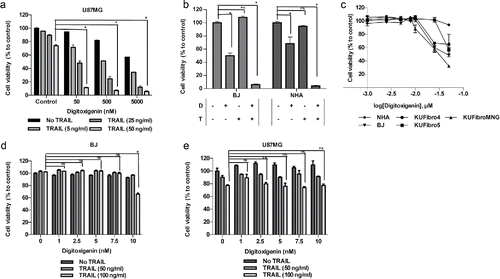
Figure 3. Effect of a DNA damaging agent, Mitoxantrone, on GBM and non-malignant cell viability (A) Viability of U87MG cells upon 0.02–20 μM of Mitoxantrone treatment. (B) The effect of different dose combinations of Mitoxantrone and TRAIL on U87MG cell viability. (C) The effects of Mitoxantrone doses between 50–5000 nM on primary human fibroblasts: KUFibro5, KUFibroMNG and established cell lines, BJ and human astrocytes (NHA). (D) The effect of the combination Mitoxantrone (500 nM) and TRAIL 50ng/ml) on BJ and NHA. (E) Live cell imaging of U87MG cells treated with Mitoxantrone, TRAIL and the combination for 24 hours. Left: graph depicting viable cell numbers, Right: representative snapshot images at t = 0 and t = 24 hours. Statistics were performed by 2-way ANOVA with Tukey's post hoc testing. (F) Caspase-8, −9 and −3/7 level activity of Mitoxantrone and/or TRAIL-treated U87MG cells. (G) Detection of apoptosis markers, cleaved PARP and cleaved caspase-3 protein levels, by Western Blot. *Indicates p <0.05, *** indicates p<0.0001 and ns stands for nonsignificant.
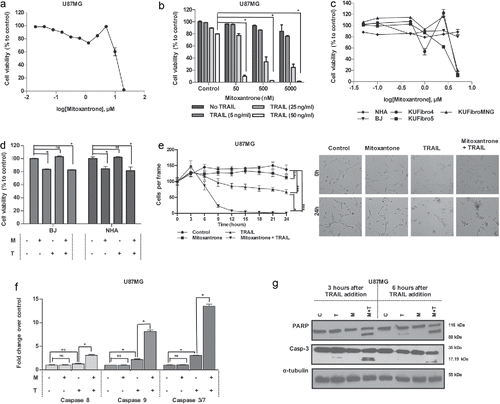
Figure 4. The effect of Mitoxantrone on TRAIL-resistant subpopulation, U87MG-R50. (A) The difference of U87MG cells and U87MG-R50 cells response to different doses (50, 250 and 500 ng/ml) of TRAIL. (B) Scatter plot of U87MG cell viability upon 24 hours of treatment with drug library composed of 1200 compounds and TRAIL together. Each dot represents the combinatorial effect of the individual drug (5 µM) and TRAIL (25 ng/ml) on cell viability compared to untreated control samples. Horizontal dashed lines depict the range of viability within 3 SD of the mean viability (41% and 113%). Red and blue dots indicate the combination effect below the threshold of 3 SD from the mean cell viability. (C) The effect of Mitoxantrone (500 nM) and/or TRAIL (50 ng/ml) on U87MG-R50 cells. (D) Detection of Caspase-8, −9 and −3/7 activity levels of Mitoxantrone and/or TRAIL treated U87MG-R50 cells. * indicates p <0.05 and ns indicates nonsignificant (p>0.05).
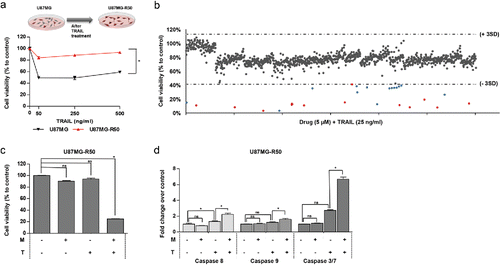
Figure 5. The effect of Mitoxantrone on TRAIL resistant cell line, U373. (A) The effect of TRAIL (5–500 ng/ml) on intrinsically TRAIL resistant cells U373. (B) Effects of 0.02–20 μM of Mitoxantrone on U373 viability. (C) The effect as single agents and of the combination of the chosen dose of Mitoxantrone and TRAIL on U373 cells. (D) Caspase-8, −9 and −3/7 level detections of Mitoxantrone and/or TRAIL treated U373 cells. (E) The effect of caspase-8 or −9 inhibitors and general caspase inhibitors on U373 cells treated with Mitoxantrone, TRAIL and the combination. * indicates p <0.05 and ns indicates nonsignificant (p>0.05).
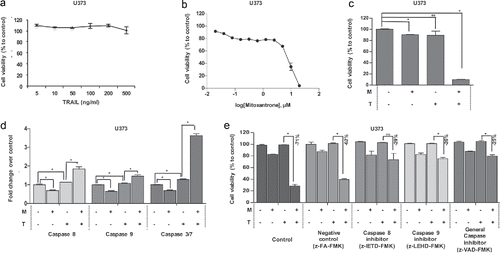
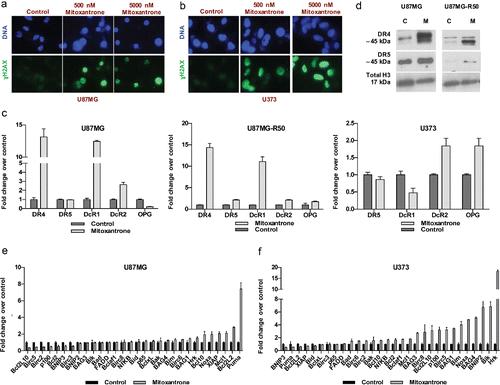
Figure 6. The effect of Mitoxantrone on apoptosis pathway. (A-B) γH2AX staining of U87MG cells (A) and U373 cells (B) treated with 0, 500 and 5000 nM Mitoxantrone. (C) Gene expression levels of death receptors of U87MG, U87MG-R50 and U373 cells treated with Mitoxantrone for 24 hours. (D) DR4 and DR5 protein levels of U87MG and U87MG-R50 cells treated with Mitoxantrone for 24 hours. (E-F) The expression of pro-apoptotic and anti-apoptotic genes of U87MG cells (E) and U373 cells (F) treated with Mitoxantrone for 24 hours.