Figures & data
Figure 1. Both normal MBD4 protein and TruMBD4 localize to the nucleus in colorectal cancer cells. (A) Upper row: light microscopy of human colon cancer cell lines HT29 (MMR-proficient, MBD4A10/A10), HCT116 (hMLH1−/-, MBD4A10/A9) and HCA7 (hMLH1−/-, MBD4A9/A9); Second row: Indirect immunofluorescence microscopy utilizing anti-MBD4 antibody with Alexa Fluor 594-conjugated secondary antibody (staining red); Third row: DAPI staining of nuclei; Fourth row: merge of second and third row images. (B) Western blot of colorectal cancer cell nuclear lysates for MBD4 and TruMBD4 expression. Signals were detected by an LAS-4000 luminescent image analyzer (GE Healthcare Bio-Sciences) utilizing a chemiluminescent solution.
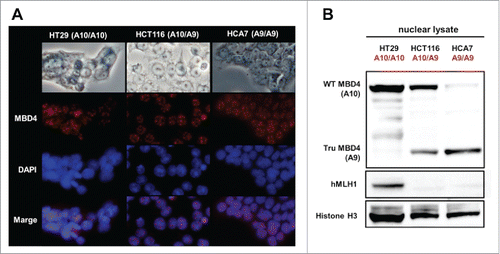
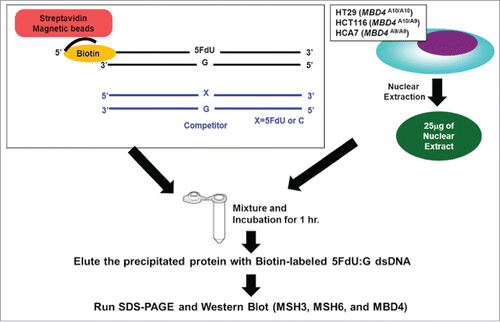
Figure 3. TruMBD4 binds to 5FU incorporated into DNA with higher affinity than normal MBD4 protein, and reduces 5FU affinity of DNA mismatch repair proteins in DNA pull down assays. (A) (Upper panel) Nuclear lysates from HT29 cells containing protein derived from wild type MBD4. As amounts of 5FdU:G competitor is increased relative to bound biotin-labeled 5FdU:G, the amount of MBD4 protein precipitated by biotin-labeled 5FdU:G is partially reduced, indicating some MBD4 protein was bound (or stolen) by the 5FdU:G competitor. This was not the case with a control complementary competitor, indicating MBD4 specifically recognizes 5FU within DNA (not shown). (Lower panel) Nuclear lysates from HT29 containing protein derived from frameshifted mutant MBD4 (TruMBD4). Here, as amounts of 5FdU:G competitor increased relative to biotin-labeled 5FdU:G, the amount of TruMBD4 precipitated by biotin-labeled 5FdU:G DNA is markedly reduced, suggesting a relative higher affinity for 5FdU:G as compared to normal MBD4 protein. (B) Bar graphs representing the reduction (competed off) rate by the protein for the 5FdU:G competitor, equating to the relative affinity level of the protein for 5FU within DNA. The affinity level of TruMBD4 for 5FdU:G is markedly higher than that of normal MBD4 protein. (C) Nuclear lysates from HCT116 cells containing both normal MBD4 protein and TruMBD4. As amounts of 5FdU:G competitor is increased, both normal MBD4 protein and TruMBD4 are competed off, but at apparently different rates. (D) Bar graph representing the competed off rate for both normal MBD4 protein and TruMBD4 from HCT116 cells. The relative affinity level of TruMBD4 was higher than normal MBD4 protein for the 5FdU:G competitor. (E, F) Nuclear lysates from HCA7 cells (E) and HT29 cells (F), demonstrating the relative pull down and competition off binding by MBD4/TruMBD4, MSH3 (key component of the hMutSβ MMR recognition complex) and MSH6 (key component of the hMutSα MMR recognition complex) for 5FdU:G. Note the relative difficulty for “compete off” bound reduction for MSH3 and MSH6 by the 5FdU:G competitor when TruMBD4 is present, compared to the “compete off” reduction for MSH3 and MSH6 when normal MBD4 protein is present. (F) Bar graph representing the competed off rate for MSH3 and MSH6 for the 5FdU:G competitor in the presence of normal MBD4 protein or TruMBD4. With normal MBD4 protein, MSH6 shows higher affinity for 5FU within DNA (as expected). However the binding affinity rates of both MMR proteins are markedly lower in TruMBD4-expressed cells than that of normal MBD4-expressed cells.
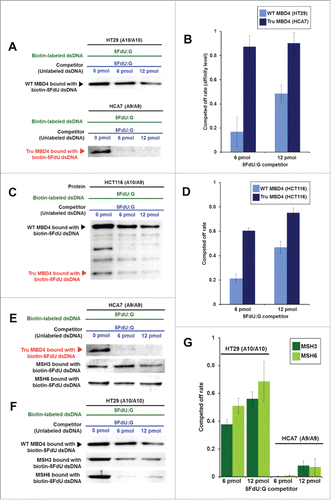
Figure 4. TruMBD4 enhances 5FU cytotoxicity in hMLH1-proficient cells. (A) Establishment of stable, TruMBD4-expressed HT29 cell clones as shown by Western blot (right lane). Cells were transfected with a pcDNA3 plasmid (Invitrogen) encoding TruMBD4, and selected by G418. HCT116 lysates served as a positive control since it expresses both normal MBD4 protein and TruMBD4 (left lane). HT29 cells transfected with an empty pcDNA3 plasmid served as negative control (middle lane). β-actin served as a loading control. (B) MTS assay. Cells were seeded at a density of 5000 cells per well into 96-well plates in culture medium treated with 5 μM, 10 μM;, 15 μM;, 20 μM of 5FU. After 5 d of growth, the number of viable cells was counted via the assay. (C) Clonogenic assay. Cells were plated in growth medium supplemented by 10% FBS and containing various concentrations of 5-FU (0, 5, and 10 µM). After 10 d of growth, the culture plates were washed, fixed with methanol, and stained with 3% Giemsa. From both MTS and clonogenic assays, TruMBD4 enhances 5FU-induced cytotoxicity.
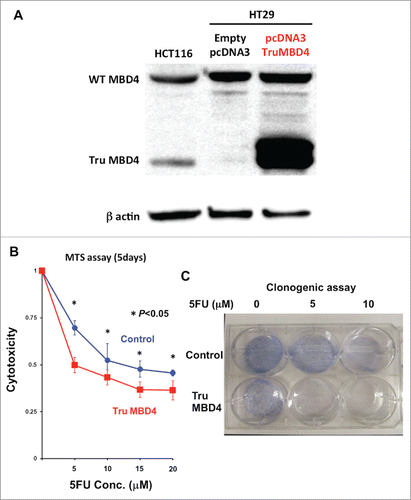
Figure 5. TruMBD4 induces S phase cell cycle arrest upon 5FU treatment. (A) Using fluorescence-activated cell sorting (FACS) analysis, empty plasmid transfected HT29 cells expressing normal MBD4 protein showed a modest increase in S phase cells after 5FU treatment (16.2% pre-treatment, 24.3% post 5FU treatment). (B) TruMBD4-expressing HT29 cells, in response to 5FU treatment, demonstrated marked increase in S phase cells (18.9% pre-5FU treatment, 55.0% post 5FU treatment).
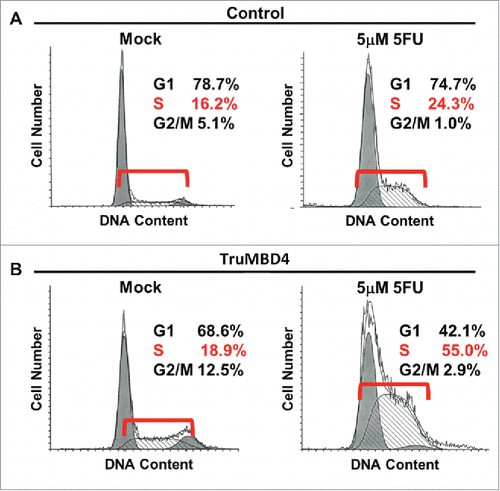
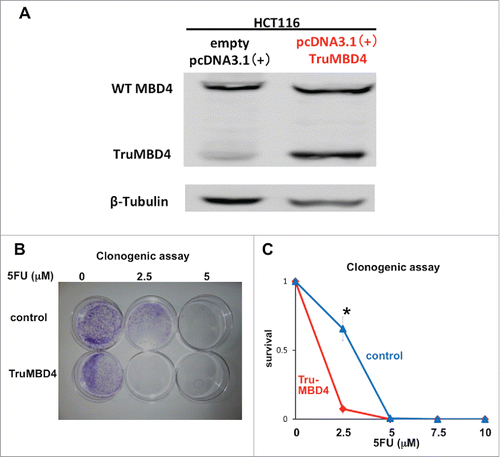
Figure 6. TruMBD4 enhances 5FU cytotoxicity independent of hMLH1 status. (A) Establishment of stable, TruMBD4-expressed HCT116 cell clones as shown by Western blot (right lane). Cells were transfected with a pcDNA3 plasmid (Invitrogen) encoding TruMBD4, and selected by G418. HCT116 cells transfected with an empty pcDNA3 plasmid served as negative control (left lane). β-actin served as a loading control. Note the marked increase in TruMBD4 expression relative to normal MBD4 protein expression. (B, C) Clonogenic assay. Cells were plated in growth medium supplemented by 10% FBS and containing various concentrations of 5-FU (0, 5, and 10 µM). After 10 d of growth, the culture plates were washed, fixed with methanol, and stained with 3% Giemsa (B). Previously viable clonal colonies of at least 50 cells were counted. The relative surviving fraction for each cell line was expressed as a ratio of the plating efficiency in treated cultures to that observed in the controls (C). The enhanced TruMBD4-overexpressed HCT116 cells increased 5FU cytotoxicity over control cells (*P<0.05 at 2.5 µM 5FU).