Figures & data
Figure 1. Effect of mir-339 and miR-766 on DNMT3B expression: HEK293T, which was transfected with the Renilla and firefly luciferase expression construct psiCHECK™-2 harboring DNMT3B 3′-UTR and pLEX-JRed-TurboGFP containing miR-339, miR-766 and miR-scrambled. After 24 h, Data represents mean ± SE from 3 independent experiments performed in triplicates vs. the control group (P < 0.05).
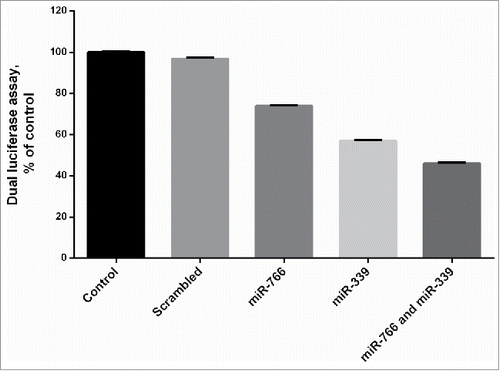
Figure 2. Expression of miR-766 and miR-339 in different cell lines before and after transduction with lenti-miR-766 and lenti-miR-339. The asterisks indicate the groups which were significantly different (p < 0.05) from each other, and the ND indicates not detectable differences between groups.
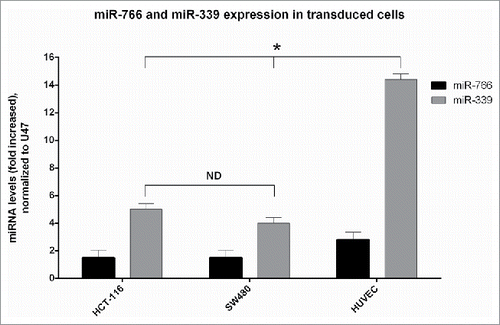
Figure 3. DNMT3B expression: SW480, HCT 116 and HUVEC were transduced with lenti-miR-339, lenti-miR-766 and lenti-miR-scrambled. After 72 hours, DNMT3B expression was analyzed by Real-Time quantitative PCR (P < 0.05).
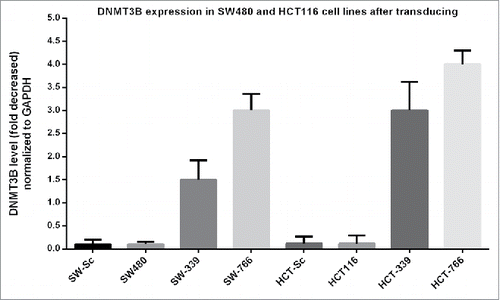
Figure 4. Graphs of some High Resolution Melting curves analysis: Some representative graphs for HRM on SFRP1, SFRP2, WIF1, DKK2, CDK2N2A, CACNA2 and hMLH1 genes beside controls. Controls composed of 0%, 25% 50% 75% and 100% methylated samples. Percentage of methylation in unknown (studied) samples calculated according to the melting curve pattern in comparison with controls. ) Decreased methylation of DKK2 gene in HCT-766 cell line to 0%. b) Methylation of DKK2 gene in SW-766 and SW-339 cell line, which is shown to be 0% and 25%, respectively. c) No alteration in methylation level of SFRP-1 gene in HCT-339 but decreased methylation in HCT-766 cell line to 0%. d) Decreased methylation of SFRP1 gene in SW-339 and SW-766 cell line, to 50% and 0%, respectively. e) Decreased methylation of SFRP2 gene to 0–25% in HCT-766 and no alteration in methylation level of HCT-339 cell line. f) Decreased methylation of SFRP2 gene to 0% and 50% in SW-766 and SW-339 cell lines, respectively. g) Decreased methylation of WIF1 gene in HCT-766 and HCT-339 cell lines to 0–25% and 25% respectively. h) Decreased methylation of WIF1 gene in SW-766 and SW-339 cell lines to 0–25% and 25%, respectively. i) The methylation level of DKK2 gene in HUVEC-766, HUVEC-339, and HUVEC-Sc and non-transduced cells (all 0%). j) Unchanged methylation level of hMlH1 gene in SW-766, SW-339, SW-Sc and SW480 cell lines.
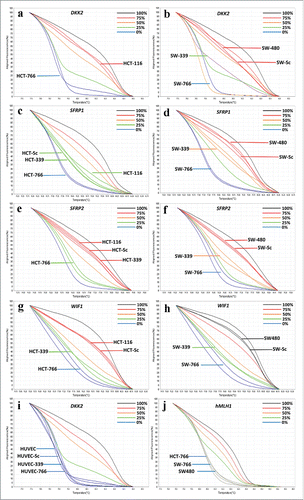
Table 1. The effect of miR-339 and miR-766 on the methylation levels of some tumor suppressor genes.
Figure 5. Expression analysis of studied tumor suppressor genes: Expression was calculated according to 2−ΔΔCt (fold change) formula compared to the data of non-transduced cells (P < 0.05). Note: the expression of all genes was not significantly different from non-transduced cells in HUVEC-766, HUVEC-339 and HUVEC-Sc.
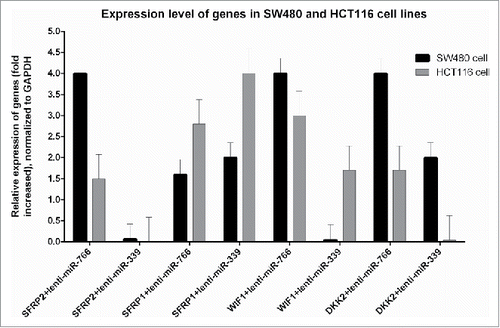
Figure 6. Results of Cell proliferation assays: Cell proliferation was evaluated by MTT proliferation assay 24 and 48 hours after transferring to a 96-well plate.
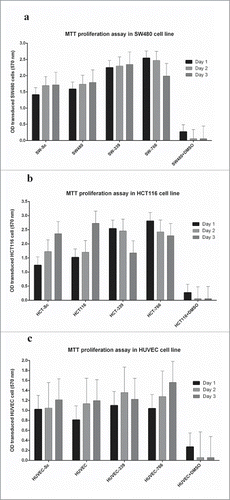
Figure 7. Cell cycle analysis in transduced and non-transduced cells: Green graphs represent the G1, blue graphs represent the s phase and the yellow graphs represent the cells in G2 phase. Transduction with lenti-miR-766(), lenti-miR-336() and lenti-miR-scrambled cells () made no changes in the cell cycle compared to non-transduced HUVEC (). Transduction of HCT116 cells by lenti-miR-766 () decreased proliferation of cells compared with HCT116 () and HCT-Sc (). On the other hand, transduction of SW480 cells by lenti-miR766 () and lenti-miR-339 () decreased G1-phase population compared to SW480 () and SW-Sc cells ().
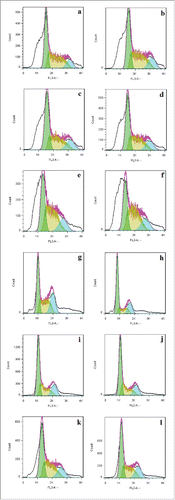
Table 3. Cell cycle statistics; distribution of cells in different phases of cell cycle after transfection with miR-766, miR-339 and miR-scrambled
Table 2. Primer sequences used for High Resolution Melting and Real-Time quantitative PCR, miRNA expression analysis and amplification of DNMT3B, 3UTR (Bold sequences indicate the endonuclease restriction site).
