Figures & data
Figure 1. (A) Hierarchical clustering of the 1,000 best discriminating genes between the standard risk C1 and the high risk C2 HB subclasses. Important clinicopathological characteristics are depicted below. A detailed list of the genes can be found in Suppl. Table 1. (B) Bar graphs represent the Connectivity Score data for vorinostat (SAHA) and trichostatin A (TSA). The black horizontal lines represent each instance performed with the respective compound. Instances in the red area indicate negative correlation scores and instances in the green area positive ones. No correlation can be detected for instances in the gray area.
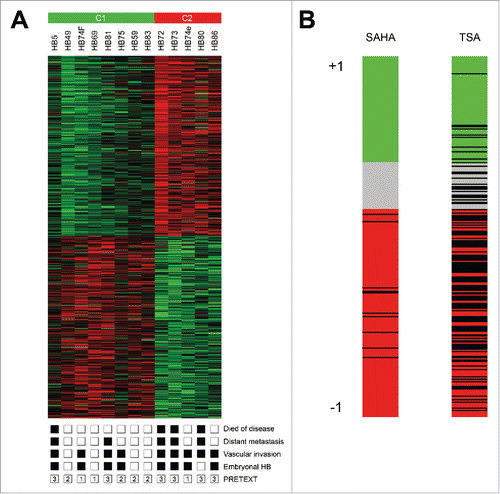
Table 1. Results of Connectivity Map analysis.
Figure 2. (A) HDAC expression levels of normal liver, primary HB and liver tumor cell lines (CL). Expression of class I and class IIa HDACs were measured by qRT-PCR and normalized to the expression of the house-keeping gene TBP. Statistical significance was calculated for differences between normal liver tissue and tumors and tumor cell lines using t-test. (B) HDAC expression levels of primary HB after their stratification as standard (C1) or high risk HB (C2) according to the 16-gene HB classifier. Statistical significance was calculated for differences between C1 and C2 tumors using t-test.
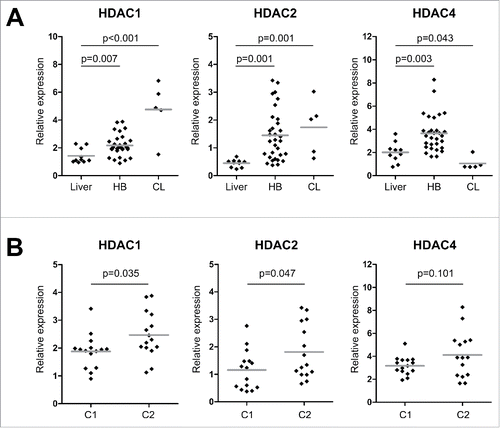
Figure 3. (A) Cell viability of HB cell lines and fibroblasts as evaluated by MTT assay after 48 h treatment with indicated concentrations of MC1568 and SAHA. Values represent means ± standard deviation of 3 independent experiments performed in duplicates. (B) Cell cycle distribution (left panel) and apoptosis (right panel) of liver tumor cell lines were analyzed by flow cytometry 48 h after treatment with vehicle (DMSO), 10µM of MC1568 and 1µM (HUH6, HepT1 and HepG2) or 2µM of SAHA (HUH7). (C) Western blot analysis for cleaved PARP in indicated HB cell lines after 48 h of treatment with DMSO, MC1568 and SAHA (concentrations as in B). (D) Expression levels of the tumor suppressor genes SFRP1, HHIP, and IGFBP3 in liver tumor cell lines after HDACi. Expression levels were measured after 48 h of treatment by qRT-PCR and normalized to the expression of the house-keeping gene TBP. Indicated are fold changes to the individual DMSO control.
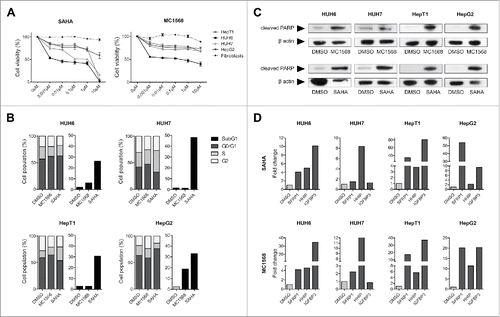
Figure 4. (A) Overall survival was calculated as time from diagnosis to death of the disease and is plotted for 30 HB patients. Statistical significance was calculated using the Mantel-Cox test. (B) Cell lines expressing the adverse C2 signature were treated with 10µM of MC1568, 1µM (HUH6, HepT1 and HepG2) and 2µM (HUH7) of SAHA, or vehicle (DMSO). Graphs show decreased expression levels in percent of 8 high risk C2 signature genes of the indicated genes upon 48 h of treatment with the indicated HDAC inhibitors.
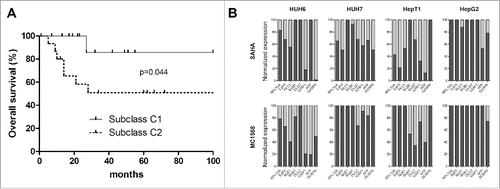
Table 2. Association between C1/C2 classification and clinicopathological features of HB.
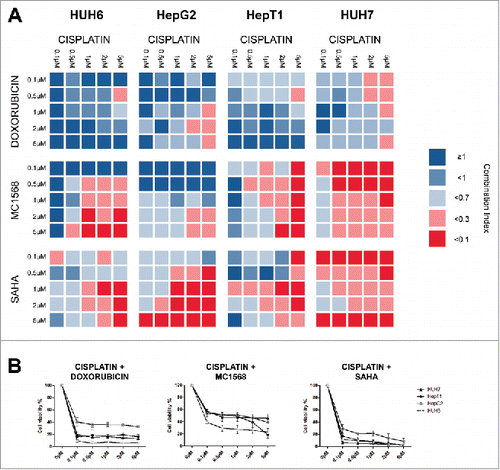
Figure 5. (A) Cell lines were treated with the indicated concentrations of cisplatin combined with various concentration of either doxorubicin, MC1568 or SAHA. Cell viability was measured after 48 h and combination indices (CI) were calculated from 2 independent experiments performed in duplets. CI < 0.1 = very strong synergism, CI < 0.3 = strong synergism, CI < 0.7 = synergism, CI <1 = slight synergism, CI ≥ 1 no synergism. (B) Cell lines were treated with the indicated concentrations of cisplatin combined with 5µM of either doxorubicin, MC1568 or SAHA. Cell viability was measured after 48 h from 2 independent experiments performed in duplets.