Figures & data
Table 1. The top 10 most enriched up and downregulated lncRNAs.
Figure 1. The top 5 pathways of GO and KEGG enrichment analysis in differentially expressed IncRNA. The horizontal axis included the score of enrichment while the vertical axis represented the enriched pathways. GO annotation of upregulated IncRNA was displayed in (A) while the downregulated ones were in (B). (C) was the KEGG annotation of upregulated lncRNA while (D) was that of downregulated lncRNA. The higher the enrichment score was, the more the pathways were involved in cancer regulation.
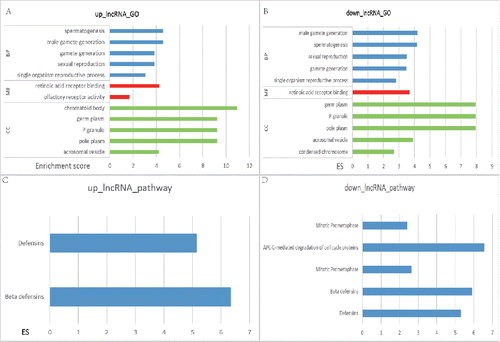
Table 2. The top 10 most enriched genes of TSGs.
Table 3. The top 10 up and downregulated oncogenes.
Figure 2. Heatmap of the top 100 differentially expressed TSGs between normal samples and tumor specimens. The horizontal axis were different samples, the gray ones were tumor samples while the yellow ones were normal specimens. The vertical axis was genes, which indicated that there was obvious difference among normal samples and tumor ones.

Figure 3. The top 100 differentially exprssed oncogenes between normal samples and tumor specimens. The horizontal axis was samples whie the gray ones were tumor samples and the yellow ones were normal specimens. The vertical axis stands for different genes.
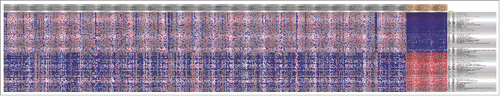
Figure 4. The most enriched data set of differentially expressed up and downregulated oncogenes. (A) represents the downregulated genes in KEGG_MAPK_SIGNALING_PATHWAY data set and (B) represented how many genes were upregulated in GO data set RESPONSE_TO_DNA_DAMAGE_STIMULUSj. (C) represents the downregulated genes in PLASMA_MEMBRANE data set and (D) represented how many genes were upregulated in MORF_AATF.
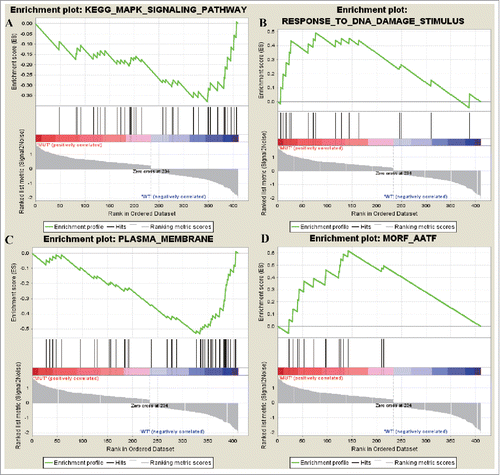
Figure 5. The most enriched data sets of tumor supressor gene. (A) represented upregulated genes in KEGG_CELL_CYCLE and (B) represented downregulated genes in KEGG_TIGHT_JUNCTION. (C) represented upregulated genes in RESPONSE_TO_DNA_DAMAGE_STIMULUS while (D) represented how many downregulated genes in PLASMA_MEMBRANE.
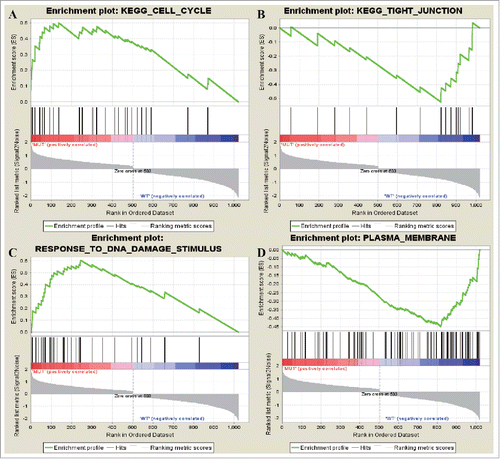
Table 4. Co-expression pairs of lncRNA and oncogenes.
Table 5. Co-expression pairs between lncRNA and TSGs.
