Figures & data
Figure 1. (a) Domains of CtBP1- Role, functions and post-translational modification sites. R, E and H signify the catalytic triad within the CtBP dehydrogenase domain (b) Graphical representation of CtBP mediated co-repression, where CtBP nucleates a complex that bridges chromatin modifiers (including histone deacteylases [HDAC] and histone methyltransferases [HMT]) to specific DNA sites recognized by a DNA-interacting protein (DNA-IP) that encodes a PxDLS domain.
![Figure 1. (a) Domains of CtBP1- Role, functions and post-translational modification sites. R, E and H signify the catalytic triad within the CtBP dehydrogenase domain (b) Graphical representation of CtBP mediated co-repression, where CtBP nucleates a complex that bridges chromatin modifiers (including histone deacteylases [HDAC] and histone methyltransferases [HMT]) to specific DNA sites recognized by a DNA-interacting protein (DNA-IP) that encodes a PxDLS domain.](/cms/asset/a0655522-19e0-4e75-b1ed-f02799cc9b33/kcbt_a_1323586_f0001_c.gif)
Figure 2. Patterns of genetic alteration of CtBP1 and CtBP2 in human solid tumors. The Cancer Genome Atlas (TCGA) Project data were extracted using cBioportal (http://www.cbioportal.org/public-portal/), and show copy number alterations or mutations in TCGA tumor collections from 15 different cancer types.
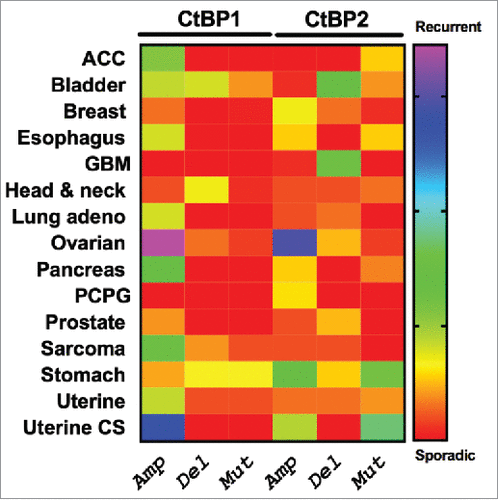
Figure 3. APC-dependent role of CtBP in the Wnt/ β-catenin pathway. In the presence of normal APC (APC-FL; left), CtBP participates in the destruction complex to sequester β-catenin (B-cat) and target it for proteasomal degradation. With mutant APC (APC-Mut/trunc; right), CtBP facilitates oligomerization and cytoplasmic sequestration of APC, promoting β-catenin release to the nucleus to induce downstream TCF/LEF mediated cell signaling, in which CtBP also plays a direct role as a coactivator at TCF target genes.Citation75,76
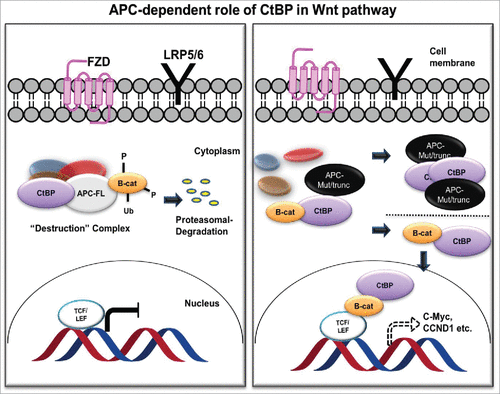
Table 1. Name and the chemical structures of CtBP inhibitors.
Figure 5. Structure of CtBP/substrate/coenzyme complex. (a) CtBP1 dehydrogenase domain structure (monomer) with domains indicated. Conformations of MTOB (light or dark blue) when interacting with (b) CtBP1 and (c) CtBP2 in presence of NAD+. (Reprinted with permission from Hilbert et al.Citation88 Copyright 2014 John Wiley & Sons, Inc.)
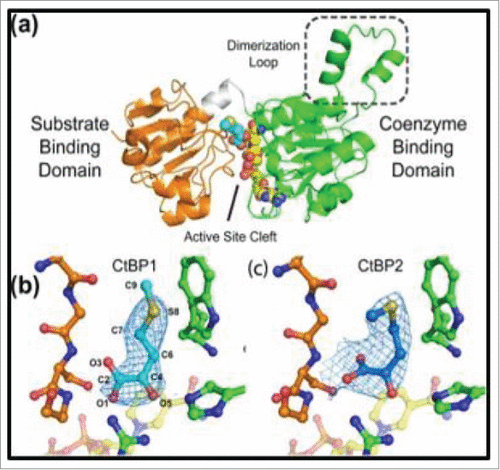
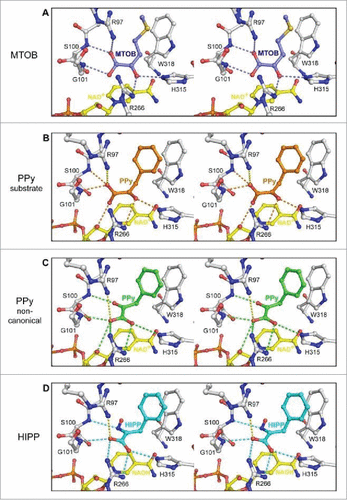
Figure 6. Ball and stick model representing interaction of CtBP1 with substrate MTOB, and substrate competitive inhibitors phenyl pyruvate (PPy) and HIPP (in stereo). (A) The hydrogen bond network of MTOB as reported in Hilbert et alCitation82 (B) Phenylpyruvate (orange) possesses a similar hydrogen bond network to MTOB (orange dashes). (C) The non-canonical phenylpyruvate conformation (green) has a distinct hydrogen bond network (green dashes). Orientation of the carboxylate toward R266 of CtBP1 maximizes hydrogen bond potential as well as coulombic interactions (yellow dashes) with R97. (D) HIPP (cyan) forms a similar hydrogen bonding network (cyan dashes) to the non-canonical phenylpyruvate conformation, with the exception of losing the interaction with the nicotinamide ribose. (Reprinted with permission from Hilbert et al.Citation85 Copyright 2015 American Chemical Society).