Figures & data
Table 1. Primer sequences used for eIF2β polylysine stretches deletion.
Figure 1. Effect of eIF2βΔ3K expression on protein synthesis. (A) Domain structure of eIF2β. Black boxes indicate polylysine stretches, dotted boxes indicate a cysteine zinc finger domain, and vertically and diagonally striped boxes indicate interpolylysine regions of eIF2β. (B) Western blot analysis of eIF2β expression after tetracycline induction. The whole protein cell extract of 105 Hek293TetR cells containing pJL (empty vector), pJL::eIF2βWT or pJL::eIF2βΔ3K were loaded into an SDS-PAGE gel and subjected to western blot analysis with anti-eIF2β and anti-TetR. The second column of each construction shows cells that were induced by tetracycline (1 µg/ml) for 72 h. The western blot was probed with antibody against tetracycline repressor protein (TetR) and used as protein loading control and to show the constitutive TetR expression in the Hek293 cell line. (C) eIF2β expression was induced by tetracycline (1 µg/mL) for 24 to 96 h in Hek293TetR cells expressing pJL, pJL::eIF2βWT or pJL::eIF2βΔ3K. Qualitative analysis of the overall protein synthesis was performed observing dEGFP expression by fluorescence microscopy. The scale bars represent 100 μm. (D) After cell sorting of Hek293TetR cells expressing pJL, pJL::eIF2βWT or pJL::eIF2βΔ3K, protein synthesis was measured using dEGFP expression in 24, 48, 72 and 96 h by flow cytometry. The anisomycin 5 µg/mL was used as a control of the protein synthesis inhibition in Hek293TetR cells transfected with pJL. The results are presented as the mean of 3 independent experiments. (E) Flow cytometry graph representative of 96 h tetracycline treatment showing the dEGFP expression shift as a consequence of eIF2βΔ3K overexpression. Purple full histogram represents untreated cells, and green line histogram represents protein synthesis induced by tetracycline (1 µg/ml).
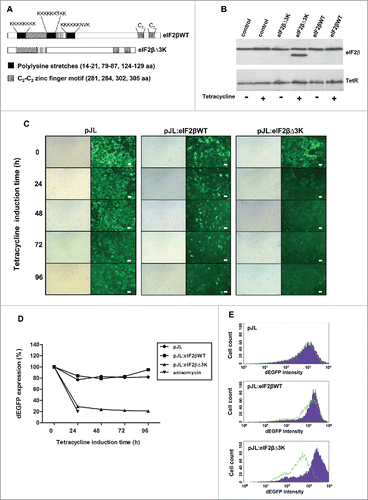
Figure 2. Western blot analysis of eIF2βWT and eIF2βΔ3K after induction with tetracycline (1 μg/mL). A shows eIF2β either endogenous and expressed from pJL::eIF2βWT, line 1: expression of eIF2βΔ3K after 24 hours of induction used as control, lines 2 to 6 expression of eIF2βWT induced after 1, 24, 48, 72 and 96 hours, respectively. B shows eIF2βΔ3K expression induced after 1, 24, 48, 72 and 96 hours, in lines 1 to 5, respectively.
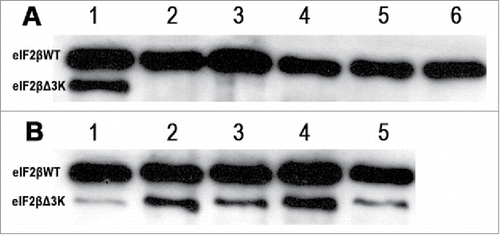
Figure 3. Effects of polylysine stretches deletion on cell viability. Gene expression of eIF2βWT or eIF2βΔ3K was induced by tetracycline (1 µg/mL) in Hek293TetR cells transfected with pJL (empty plasmid), pJL::eIF2βWT or pJL::eIF2βΔ3K. (A) Cell viability was analyzed using XTT assay after 24 to 96 h of tetracycline treatment. The viability results are expressed as the percentage (mean ± standard error mean, SEM) relative to untreated cells containing the corresponding construct. (B) Cell death was analyzed using Trypan blue exclusion method after 48 and 96 h of tetracycline induction. The death results are expressed as the percentage (mean ± SEM) of non-viable cells for each construct. Results are presented as the mean of 3 independent experiments. *indicates P < 0.05 and **indicates P < 0.01.
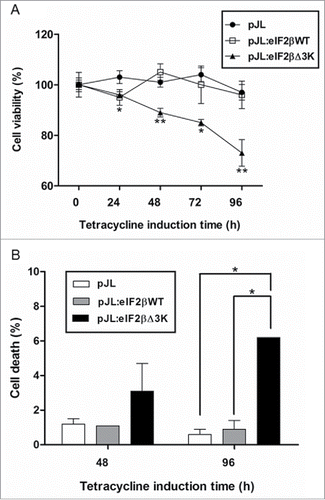
Figure 4. Deletion of the polylysine stretches affects cell proliferation. Gene expression was induced by tetracycline (1 µg/mL) in Hek293TetR cells containing pJL (empty plasmid), pJL::eIF2βWT or pJL::eIF2βΔ3K. (A) Cells were selected by cell sorting to obtain similar dEGFP expression range. Proliferation of the selected cells was analyzed by (methyl-3H)thymidine incorporation after 24, 48 and 96 h of tetracycline induction. The proliferation results are expressed as the percentage (mean ± SEM) of (methyl-3H)thymidine incorporation relative to empty plasmid at each time point. (B) Cumulative population doublings of cells were measured after 48 to 144 h of tetracycline induction. The results are expressed as the percentage (mean ± SEM) relative to tetracycline untreated cells of the corresponding construct. Results are presented as the mean of 3 independent experiments. *indicates P < 0.05, **indicates P < 0.01 and ***indicates P < 0.001.
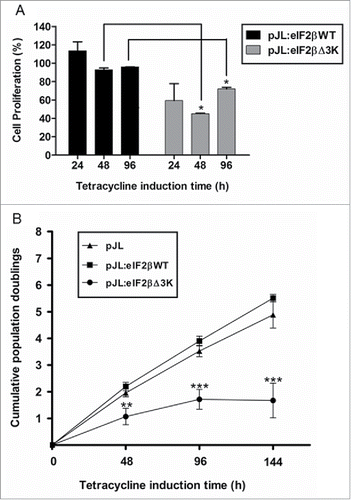
Figure 5. Effect of eIF2βΔ3K expression on cell cycle. Gene expression was induced by tetracycline (1 µg/mL) in Hek293TetR cells containing pJL (empty plasmid), pJL::eIF2βWT or pJL::eIF2βΔ3K for 96h. Cell cycle analysis was performed by flow cytometer thought PI determined DNA content and analyzed in 96h with or without tetracycline treatment. The results are expressed as the percentage of cells in different cell cycle phase. Results are presented as the mean of 3 independent experiments. Tet = tetracycline treatment.
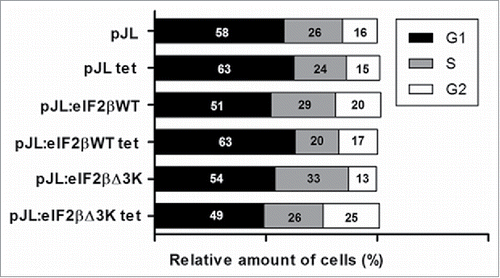
Figure 6. Subcellular localization of eIF2βWT and eIF2βΔ3K in Hek293 cell lines. Cells were transfected with empty vector pEGFP-C1 (n = 73) or plasmids carrying the fusion proteins pEGFP::eIF2βWT (n = 210) or pEGFP::eIF2βΔ3K (n = 173). Twenty-four hours after transfection, cells were fixed and submitted to immunocytochemistry using anti-nucleolus human serum. DAPI was used to stain the nucleus. The subcellular localization was analyzed by confocal microscopy. Arrow shows eIF2β nucleolar staining. (n) is the number of observed cells. The scale bars represent 10 µm.
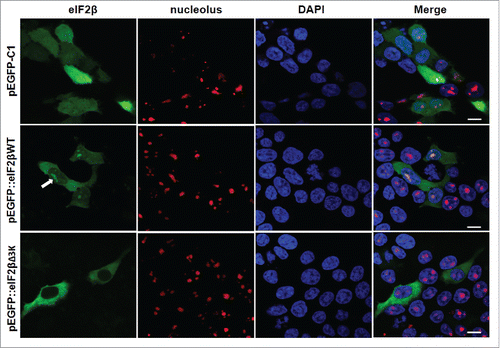
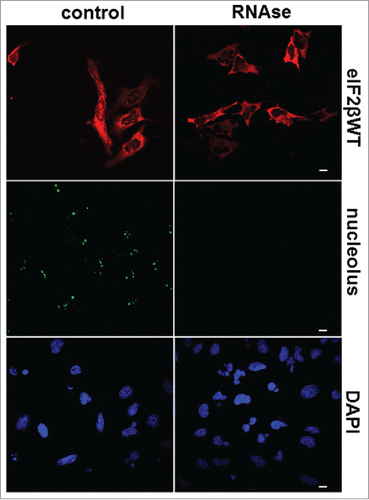
Figure 7. RNA is required for nucleolar localization of eIF2β. HeLa cells were grown on microscopic slides and transfected with pGDS::eIF2βWT. Twenty-four hours after transfection, cells were fixed with methanol (5 min, −20°C) followed by acetone (2 min, −20°C) and permeabilized with 0.1% Triton X-100 for 3 min on ice. The digestion with 0.4 mg/ml RNase A was performed in PBS for 60 min at 37°C. The cells were washed in PBS and processed for immunocytochemistry using moAb anti-eIF2β sc-133209. Control = buffer without RNase A. The anti-nucleolus human serum was used to monitor the efficiency of RNase A digestion. DAPI was used to stain DNA. The scale bars represent 10 μm.