Figures & data
Figure 1. Elaiophylin inhibited cell viability and colony formation in MM cell lines. (A) Cell viability of U266, RPMI8226, KMS11 and H929 cells after 24- and 48-h exposure to various concentrations of elaiophylin (0∼1.5 μM). (B) Effect of bortezomib on the viability of 4 MM cell lines (U266, RPMI8226, KMS11 and H929) for 48 h. (C) Colony formation assay of all cell lines with elaiophylin treatment (1.0 μM) for 14 d. Representative images from the surviving colonies in culture dishes are shown left of (D) the quantification and statistical analysis of the colonies. All data are expressed as the means ± SD for triplicate samples and were analyzed with an unpaired t-test. (*P < 0.05, **P < 0.01, and ***P < 0.001 vs. control).
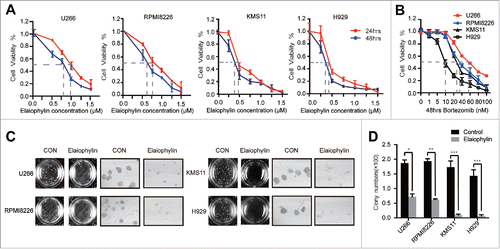
Figure 2. Elaiophylin affected cell proliferation and apoptosis. (A and B) The percentage of Edu-positive U266 and RPMI8226 cells was detected via flow cytometry after 24 h of elaiophylin treatment. (C) Flow cytometric analyses of apoptotic effects using an Annexin V-FITC/PtdIns kit. Both cell lines were incubated with elaiophylin (0∼1.5 μM) or DMSO as the control for 24 or 48 h. (D) The expression of cleaved CASP3 and cleaved PARP in 24 h was determined using western blotting. GAPDH served as a loading control. (E) CASP3 enzymatic activity in elaiophylin-treated cells for 24 h is presented as the fold change normalized to that of cells in the DMSO group. The graphs illustrate the means ± SD of 3 independent results, and the data were analyzed via ANOVA (*P < 0.05, **P < 0.01, and ***P < 0.001 vs. control).
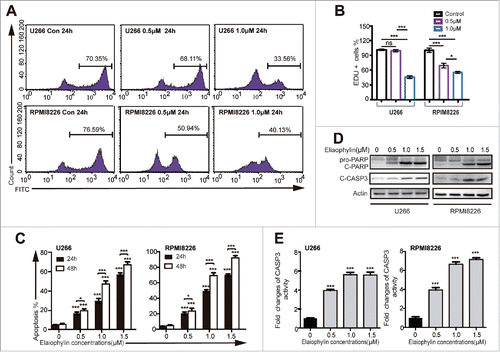
Figure 3. Elaiophylin blocked autophagy flux. (A and B) Immunofluorescence microscopy was used to detect endogenous MAP1LC3B-II puncta formation per cell. The data are presented as the means ± SD of 3 samples, with at least 100 cells in every sample (*P < 0.05, **P < 0.01, and ***P < 0.001). (C) Representative fluorescence images and (D) statistical analysis of U266 cells transfected with mRFP-GFP-LC3 lentivirus and treated with rapamycin (250 nM), chloroquine (20 μM) or elaiophylin (0.75 μM) for 12 h. (E) Conversion of MAP1LC3B-I to MAP1LC3B-II was detected via western blot in U266 cells treated with elaiophylin (0.75 μM) in the presence or absence of CQ (20 μM) for 12 h. (F) Quantification of the protein expression is shown as representative column graphs. (G) The protein expression of MAP1LC3B and SQSTM1 was determined with western blotting. RPMI8226 and U266 cells were treated with gradient concentrations of elaiophylin (0.5, 1.0 and 2.0 μM) for 24 h (upper panel) and incubated with 0.75 μM elaiophylin for the indicated time points (lower panel). For all western blots, GAPDH served as a protein loading control. Data (3 independent experiments) are expressed as the means ± SD and were analyzed via ANOVA (*P < 0.05, **P < 0.01, and ***P < 0.001 vs. control).
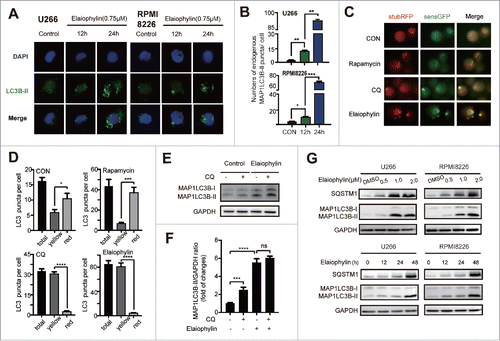
Figure 4. Elaiophylin sequentially induced ER stress. (A) Assessment of 78-kDa glucose-regulated protein (GRP78), C/EBP homologous protein (CHOP) and activating transcription factor 4 (ATF4) expression using real-time PCR in cells treated with elaiophylin (0.75 μM) for 18 and 36 h and (B) protein expression of GRP78, phospho-PERK (Thr980), ATF4, CHOP and XBP1s detected with western blotting. (C) Western blotting analysis of sequential protein expression changes in GRP78 and MAP1LC3B in cells incubated with elaiophylin (0.75 μM) for successive time points (0, 3, 6, 12, 24 and 36 h). (D) Quantification of the western blots is shown as representative column graphs. GAPDH served as a loading control. The data are shown as the means ± SD (3 independent experiments) and were analyzed using ANOVA (*P < 0.05 vs. 0 h).
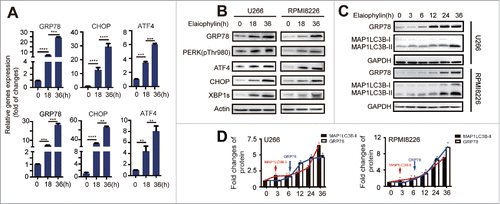
Figure 5. Elaiophylin-induced cell death partially involved ER stress-mediated apoptosis in TP53 mutant MM cell lines. U266 and RPMI8226 cells were cultured with elaiophylin (1.0 μM) or DMSO in the presence or absence of tauroursodeoxycholic acid (TUDCA, 0.75 mM) for 24 h. (A, B and C) FACS analysis of cell apoptosis. The data are presented as the means ± SD of triplicate experiments (**P < 0.01). (D) The protein expression of cleaved PARP, cleaved CASP3, GRP78 and CHOP determined by immunoblotting. GAPDH served as a loading control.
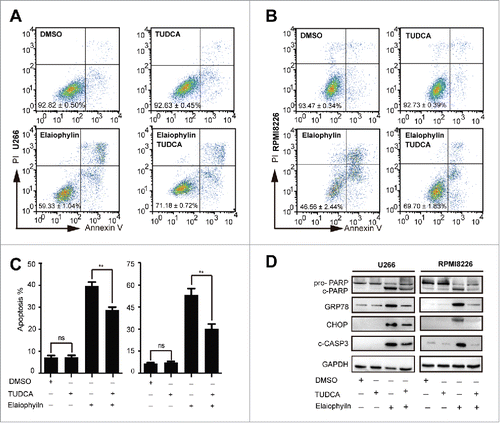
Figure 6. Elaiophylin suppressed tumor growth in a U266 xenograft zebrafish model. (A) Flowchart representing the procedures for zebrafish embryo MM xenograft model establishment and the design of the experimental grouping (G1: MM+DMSO, G2: MM+10 nM bortezomib, G3: MM + 50 μM elaiophylin, G4: wild-type + 50 μM elaiophylin, G5: wild-type + E3 buffer). (B) Fluorescence microscopy images of the CM-Dil-labeled U266 MM cells injected into the perivitelline space of zebrafish larvae (Danio rerio). (C) Representative fluorescence microscopy images of the CM-Dil-stained U266 cell xenograft growth with different treatments (Control: DMSO, bortezomib: 10 nM, elaiophylin: 50 μM) of zebrafish larvae in the perivitelline area at 0, 12, 36 and 72 h post treatment (hpt). (D) Growth curves of tumor xenografts in 3 groups (G1∼G3, n = 30 for each group). (E) Percentage of zebrafish larvae without response in the 3 groups (G1∼G3, n = 30 for each group). (F) Cumulative survival rate of zebrafish embryo xenografts 72 hpt in 5 different groups.
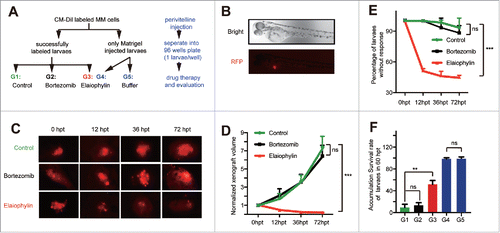
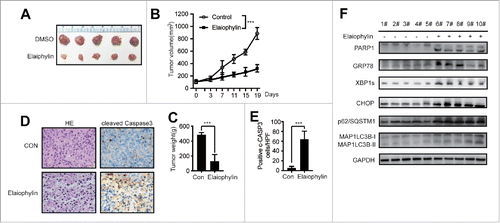
Figure 7. Elaiophylin effectively inhibited MM cells in a xenograft NOD/SCID mouse model. (A) Photographs of tumors isolated from mice after approximately 3 weeks of treatment with elaiophylin or vehicle. (B) Growth curves of the xenografts in NOD/SCID mice. (C) The statistical analysis of tumor gross weight. (D) Representative images of H&E staining (left panel) and immunohistochemical staining for cleaved CASP3 (right panel) in the tumor tissues from 2 groups. (E) Quantitation of the number of cleaved CASP3-positive cells under high magnification in 8 individual fields. (F) Western blot analyses of related protein expression in samples from the xenograft tumors in 2 groups. GAPDH was used as a loading control.