Figures & data
Figure 1. Multispectral IHC stained eight proteins involved in TGF-β signaling on TMA slides of colorectal cancer (CRC). A. Representative images for SMAD4 and p-SMAD1/5/9 staining in cancer tissues (upper) and paired normal tissues (bottom). The upper images showing the raw scanned images. B. Triple staining shows TGF-β1, SMAD2/3, and TGFBRI in cancer tissues (upper) and paired normal tissues (bottom). C. Images show the triples staining for SMAD2/3, SMAD1/5/9 and TGFBRII in cancer tissues (upper) and paired normal tissues (bottom). Nuclei were counterstained with DAPI.
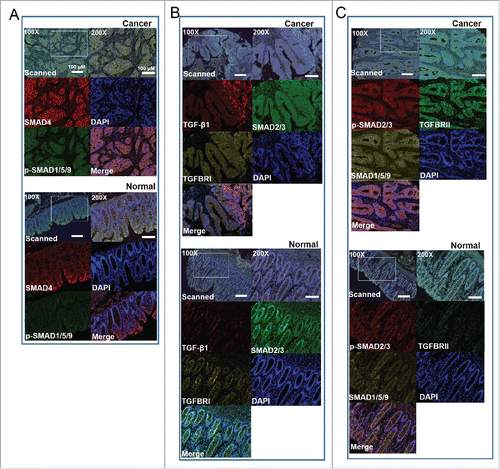
Figure 2. Subcellular locations of TGF-β signaling proteins. A. SMAD4 and p-SMAD1/5/9. B. TGF-β1, SMAD2/3, TGFBRI. C.p-SMAD2/3, SMAD1/5/9, TGFBRII.
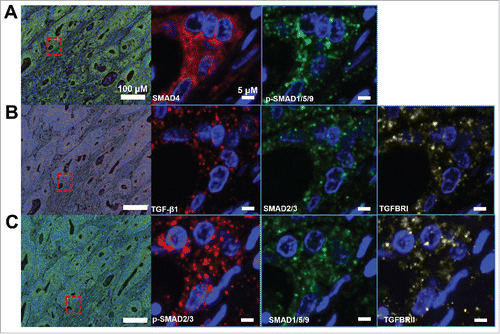
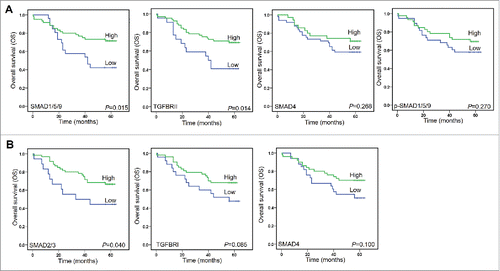
Figure 3. Scoring the stained proteins. A. Pipeline for tissues and cells segment by InForm software. The image of raw multiple fluorescent channels merged was separated into two fluorescent channels (Opal670 and DAPI) (upper). Segment tissue was divided into cancer and stroma based on the fluorescent signal intensity of DAPI (middle panel). Representative image of 0–3+(4-bin) scoring system (bottom). This score system can be used to calculate H-score with cell stains. Difference colors displays four levels (0∼1, 1∼2, 2∼3, 3∼) and percentage positivity of cells with each bin of stained protein. H-score was automatic calculated by InForm software. B. Unsupervised hierarchical clustering of eight TGF-β signaling proteins classified the 90 patients with CRC.
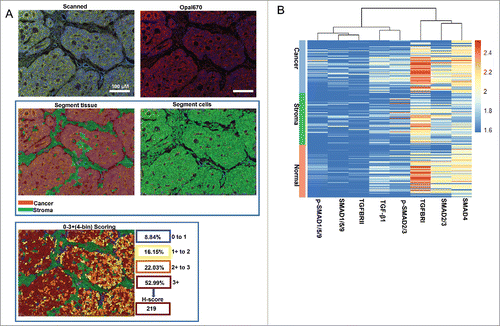
Figure 4. Comparing levels of TGF-β1 (A), TGFBRI (B), TGFBRII (C), SMAD4 (D), SMAD1/5/9 (E), p-SMAD1/5/9 (F), SMAD2/3 (G), and p-SMAD2/3 (H) based on H-Scores in cancer tissues and normal tissues of patients with CRC.
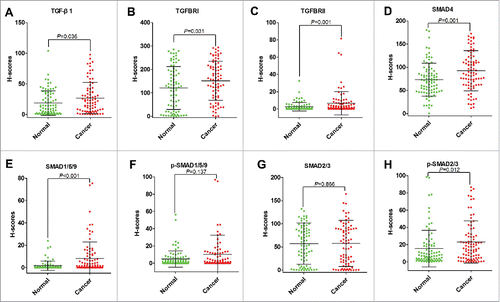
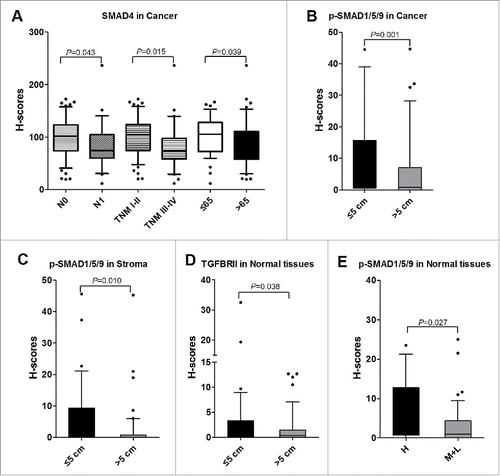
Figure 5. Correlations between TGF-β signaling and clinicopathological characteristics. A. The levels of SMAD4 associated with N stage, TNM stage, and age (years) in cancer tissues. B. The levels of p-SMAD1/5/9 were increased in CRC patients with tumor volume ≤ 5 cm than those whom tumor volume > 5cm in cancer tissues. C. In stroma, the levels of p-SMAD1/5/9 were increased in CRC patients with tumor volume ≤ 5 cm than those whom tumor volume > 5 cm. D. In normal tissues, TGFBRII was correlated with tumor volume. E. p-SMAD1/5/9 was associated with differentiation in normal tissues. H, High; M, Moderate; L, Low.