Figures & data
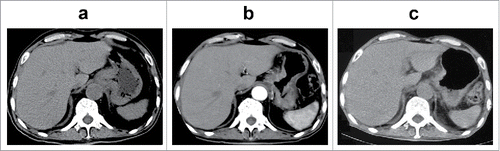
Figure 1. Imaging and histopathologic characteristics of the patient at the first diagnosis. (a and b) Positron emission tomography/computed tomography (PET-CT) scans showing heterogeneous fluorodeoxyglucose (FDG) uptake in multiple lymphadenopathies and gastric lesions. (c and d) Hematoxylin and eosin staining of biopsy tissues of the left neck lymph nodes confirmed metastatic adenocarcinoma (400 × magnification and 100 × magnification, respectively).
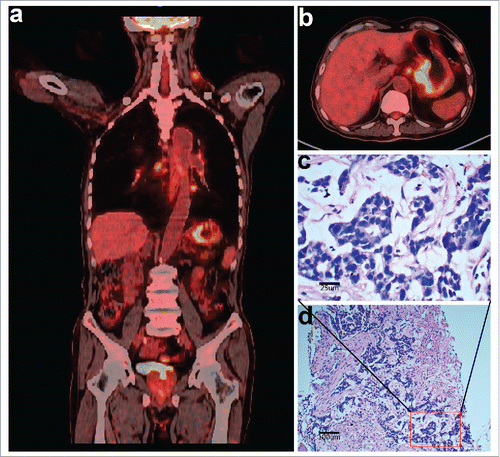
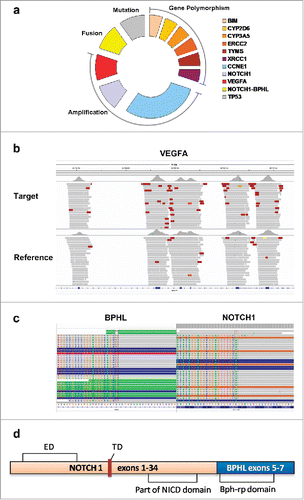
Figure 2. Molecular gene variations of the patient using targeted NGS-based tumor DNA profiling. Sequencing library was prepared by Illumina TruSeq DNA PCR-Free Sample Preparation Kit and target capture was sequenced on Illumina MiSeq NGS platform according to its instruction. Copy number variations (CNVs) were identified using tested sample and normal human hapmap DNA NA18535 average read depths at each captured region (exonic region). (a) Different forms of variation for the indicated reporters were shown. (b) VEGFA gene amplification as demonstrated by CNVs detection. (c) Paired-end sequencing data indicated somatic interchromosomal NOTCH1-BPHL fusion as demonstrated by Integrative Genomics Viewer program. (d) Schematic diagram of predicted domains of NOTCH1-BPHL fusion protein. ED: Extracellular domain; NICD: NOTCH1 intracellular domain; TD: Transmembrane domain.