Figures & data
Figure 1. Lamc1 is significantly increased in tumors of HCC patients and associated with PKM2 expression Forty tumor and 20 adjacent normal tissues of HCC patients were collected. (a,b). After isolation of RNA, the expression of Lamc1 (a) and PKM2 (b) relative to GAPDH was detected by RT-PCR. (c) The correlation between Lamc1 and PKM2 was analyzed by Pearson’s analysis (P < 0.0001).
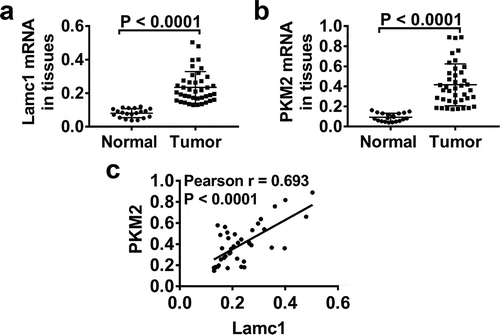
Figure 2. High expression levels of Lamc1 and PKM2 in HCC lines RT-PCR and western blot were used to detect the expression of Lamc1 and PKM2 in LO2 (normal hepatocytes), HepG2, Hep3B and Huh7 (HCC cell lines) cells. (a,b) After the RNA extraction, the mRNA expression of Lamc1 (a) and PKM2 (b) relative to GAPDH were respectively detected. (c) After the protein extraction, the levels of Lamc1 and PKM2 protein were detected. With three repeated independent experiments, the data were expressed as mean ± SD, LO2 as a control, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
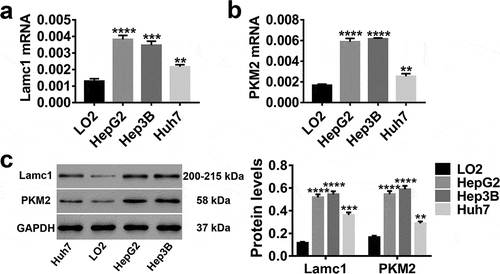
Figure 3. Down- and up-regulation of Lamc1 in HCC cell lines by lentivirus infection. (a–c) After 48 h infection of lentiviruses siLamc1 and oeLamc1, the efficiency of siLamc1 or oeLamc1 regulating Lamc1 expression in HCC cells was determined by the assays of RT-PCR (upper) and western blot (lower). With three repeated independent experiments, the data were shown as mean ± SD, **P < 0.01, ***P < 0.001, and ****P < 0.0001 compared with Vector.
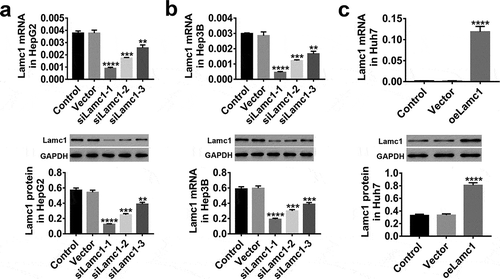
Figure 4. Lamc1 is required for the proliferation of HCC cells. After infection with siLamc1 or oeLamc1 lentivirus, (a–c) the proliferation of HCC cells was evaluated by CCK8 assays at 0, 24, 48 and 72 h. (d–f). The cell death rates of HCC cells were evaluated by Trypan blue staining. With three repeated independent experiments, the data were shown as mean ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001 compared with Vector.
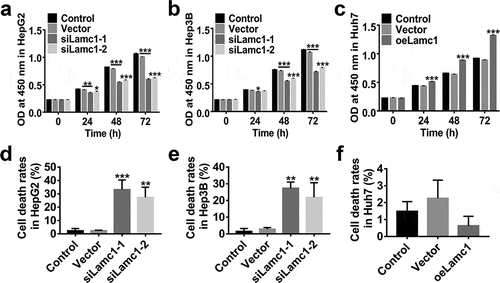
Figure 5. Down-regulation of Lamc1 inhibits the Warburg effect in HCC cells. After down- and up-regulation of Lamc1, through biochemical detections, glucose consumption (a,d,g) and lactate production (b,e,h) of these cells were measured. Percentage of glucose consumption is relative to Control. (c,f,i). The protein levels of GLUT1 and LDHA was detected by western blot. With at least three independent experiments, the data were presented as mean ± SD, **P < 0.01, and ***P < 0.001 compared with Vector.
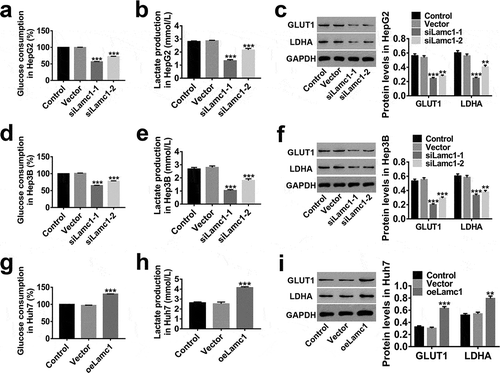
Figure 6. Lamc1 suppresses the cell proliferation and Warburg effect via regulating PKM2 (a,b). The efficiency of oePKM2 regulating PKM2 expression in HepG2 cells was determined by RT-PCR (a) and western blot (b). After co-infection of lentiviruses siLamc1 and oePKM2 in HCC cells, (c) the cell proliferation were assessed by CCK8 assays at 0, 24, 48 and 72 h. (d) The cell death rates in HepG2 were evaluated by Trypan blue staining. (e,f) The glucose consumption (e) and lactate production (f) of HCC cells were detected by biochemical detections. (g) The protein levels of PKM2, GLUT1 and LDHA were detected by western blot. With at least three independent experiments, the data were expressed as mean ± SD, **P < 0.01 and ***P < 0.001 compared with Vector, ##P < 0.01 and ###P < 0.001 compared with siLamc1.
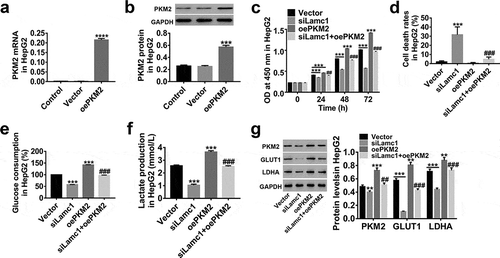
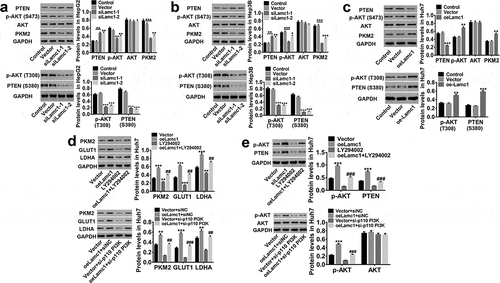
Figure 7. Lamc1 regulates PKM2 expression through the phosphorylation of PTEN/AKT pathway. (a/b/c) After Lamc1 down- and up-regulation, the protein levels of PTEN, p-PTEN (S380), p-AKT (S473/T308), AKT, PKM2 in HepG2 (a), Hep3B (b) and Huh7 (c) cells were detected by western blot analysis. (d/e) After treatment of oeLamc1 lentivirus and LY294002 or a si-p110 PI3K, the protein levels of PKM2, GLUT1, and LDHA, as well as p-AKT (T308) and AKT were detected. With three independent experiments, the data were shown as mean ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001 compared with Vector, ##P < 0.01 and ###P < 0.001 compared with oeLamc1.