Figures & data
Figure 1. LINC00963 is highly expressed in OS and associated with poor outcomes in OS patients. (a) LINC00963 RNA expression levels were assessed quantitatively in 68 pairs of osteosarcoma tissues and corresponding normal tissues utilizing qRT-PCR. (b) LINC00963 RNA expression levels were examined quantitatively in human osteoblast (hFOB) and osteosarcoma (U2OS, SAOS-2, HOS and MG63) cell lines. (c) OS patients with higher LINC00963 had a poor overall survival (N = 68, median survival:34 months vs. 46 months, Long Rank p< 0.001). (d) OS patients with higher LINC00963 had poor disease-free survival (N = 68, median survival:28 months vs. 42 months, Long Rank p < 0.001). Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
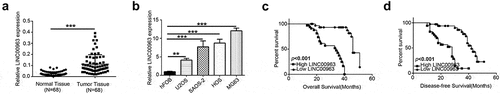
Figure 2. LINC00963 enhances proliferation and migration of OS cells. (a) The transfection effect of LINC00963 overexpression was determined by qRT-PCR. (b) The transfection effect of LINC00963 underexpression was determined by qRT-PCR. (c) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 overexpression. (d) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 underexpression. (e) Wound healing assays were performed to assess cell mobility after LINC00963 overexpression. (f) Wound healing assays were performed to assess cell mobility after LINC00963 underexpression. (g) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 overexpression. (h) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 underexpression. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
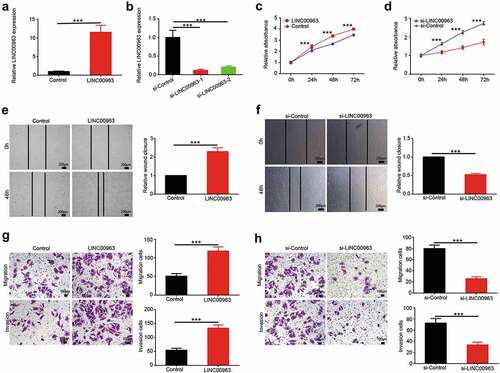
Figure 3. LINC00963 downregulates miR-204-3p expression and negative correlation between LINC00963 RNA and miR-204-3p RNA expression. (a) Potential binding targets were predicted on LncBase Predicted v.2. (b) Predicted sequence of LINC00963 wild-type 3ʹ-UTR was predicted to bind with miR-204-3p and its corresponding LINC00963 mutant 3ʹ-UTR. (c) Luciferase activity assay showed that LINC00963 can bind directly with miR-204-3p. (d) miR-204-3p RNA expression levels were measured after LINC00963 overexpression or knockdown in OS cells. (e)miR-204-3p RNA expression levels were assessed quantitatively in 68 pairs of osteosarcoma tissues and corresponding normal tissues utilizing qRT-PCR. (f) LINC00963 RNA expression levels were inversely correlated with miR-204-3p RNA expression levels in 68 osteosarcoma tissues. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).
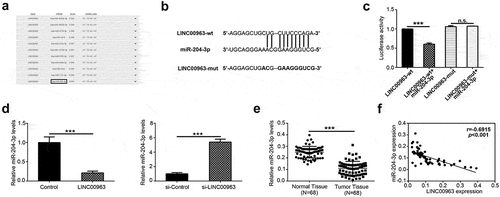
Figure 4. miR-204-3p attenuates the effects of LINC00963 on osteosarcoma cells. (a) miR-204-3p RNA expression level was examined after LINC00963 or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (b) miR-204-3p RNA expression level was examined after si-LINC00963 or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (c) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (d) Cell counting kit-8 (CCK8) assays were carried out to investigate cell growth after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (e) Wound healing assays were performed to assess cell mobility after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (f) Wound healing assays were performed to assess cell mobility after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. (g) Transwell migration and invasion assays were conducted to examine cell migration and invasion after LINC00963 or miR-204-3p mimics or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells. (h) Transwell migration and invasion assays were conducted to examine cell migration and invasion after si-LINC00963 or miR-204-3p inhibitors or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance). NC:LINC00963 control, si-NC:si-LINC00963 control, Lnc: LINC00963, si-Lnc: si-LINC00963, miR-MI: miR-204-3p mimics, miR-IN: miR-204-3p inhibitors, MI-Ctrl: miR-204-3p mimics control, IN-Ctrl: miR-204-3p inhibitors control.
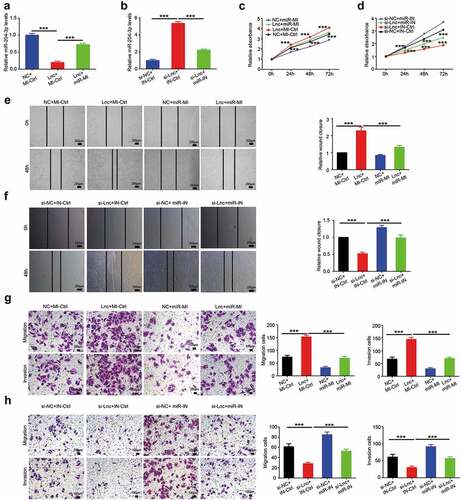
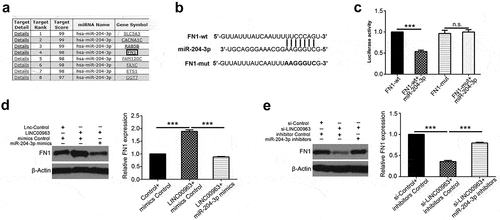
Figure 5. LINC00963 increases FN1 expression by suppressing miR-204-3p in OS. (a) Potential binding targets were predicted on miRDB. (b) Predicted sequence of FN1 wild-type 3ʹ-UTR was predicted to bind with miR-204-3p and its corresponding FN1 mutant 3ʹ-UTR. (c) Luciferase activity assay showed that FN1 can bind directly with miR-204-3p. (d) FN1 protein level was examined after LINC00963 or LINC00963+ miR-204-3p mimics or control was transfected in MG63 cells and after si-LINC00963 or si-LINC00963+ miR-204-3p inhibitors or control was transfected in U2OS cells. Data are shown Mean ± SD, *P < 0.05, **P < 0.01, *** P < 0.001, n.s. (no significance).