Figures & data
Table 1. Enrichment data for the hCD16a-VHH library.
Figure 1. Anti-hCD16a single domain antibody screening. ELISA assay was performed using 0.5 µg of purified and quantified phage. OD450 = 1.25 was used as cutoff to select out 6 clones (black bars).
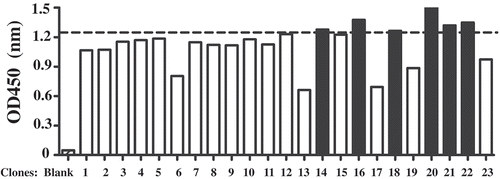
Figure 2. Biochemical characterization of anti CEA-CD16a VHHs. (a) The anti CEA-CD16a VHH SBC74-79 were constructed by fusing an anti-CEA and anti-CD16a single domain antibodies with a (GGGGS)3 linker. A His-tag was added to the C-terminal end to protein detection and purification. (b) Coomassie blue staining of purified proteins (SBC74, SBC75, SBC76, SBC77, SBC78, SBC79) after Ni-NTA affinity chromatography. (c) Gel filtration of protein marker (top panel) and SBC77 (bottom panel). (d) DLS experiment was performed as described in the Materials and Methods using 0.5 mg/ml of purified protein from 25°C–75°C.
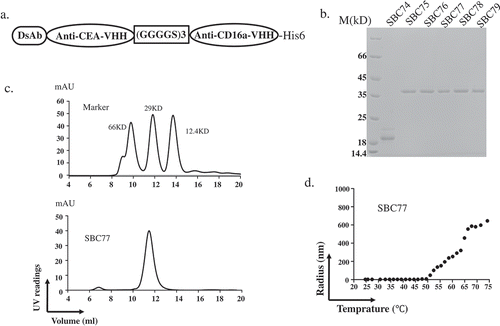
Figure 3. Anti CEA-CD16a bispecific antibodies can bind CEA and CD16a antigen. a) Flow cytometry analysis of anti CEA-CD16a bispecific antibodies were performed using LS174T and SKOV3 cells. Light gray area (blank), indicating LS174T cells with no staining; Dark gray area (blank), indicating SKOV3 cells with no staining; dotted line, cells with anti-His-PE staining; Solid line, anti CEA-CD16a VHHs and then anti-His-PE staining. b) Elisa analysis of different bispecific antibodies binding to CD16a antigen, BiSS (gray, dashed line); Anti CEA-CD16a bispecfic antibodies SBC75-79 (black, solid line); No CD16a(gray, solid line). The data are the mean of triplicates with error bars representing the standard deviation.
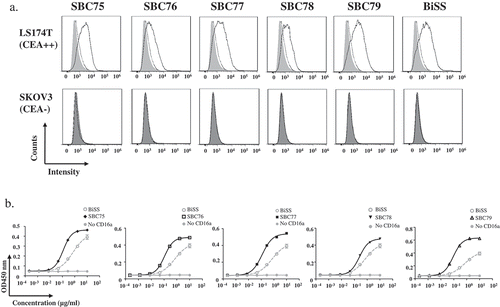
Figure 4. Anti CEA-CD16a bispecific antibodies induce NK cell-mediated cytotoxicity. a) Different cell lines were treated with anti CEA-CD16a bispecific antibodies or anti-CD16a VHHs with fresh isolated NK cells. The effector (NK cells) (25000 cells/well) and target cells LS174T and SKOV3 (2500 cells/well) at ratio of 10:1. All data are the mean of triplicates with error bars representing the standard deviation. (***P < .001, t test, compared with Anti-CD16a VHHs).
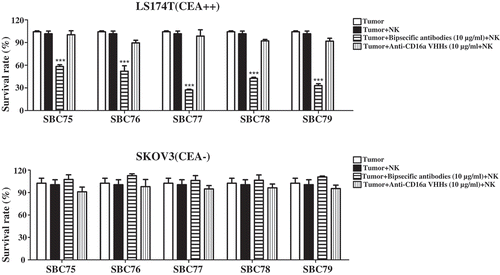
Figure 5. Anti CEA-CD16a bispecific antibodies mediates NK cell-dependent tumor cell killing. (a) Different cell lines were treated with anti CEA-CD16a bispecific antibodies with or without fresh isolated NK cells. The effector (NK cells) (25000 cells/well) and target cells LS174T, HT29 and SKOV3 (2500 cells/well) at ratio of 10:1. All data are the mean of triplicates with error bars representing the standard deviation. (***P < .001, t test, compared with Tumor+NK). (b) SBC77 induce NK cell-mediated cytotoxicity in a dose dependent manner. The concentration of BiSS, SBC77 and Anti-CD16a VHH (D5) are from 0.001 ng/mL to 10 µg/mL. All data are the mean of triplicates with error bars representing the standard deviation.
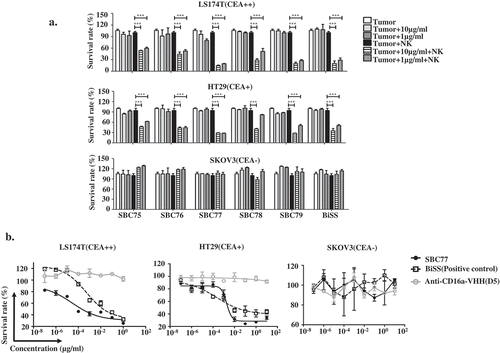
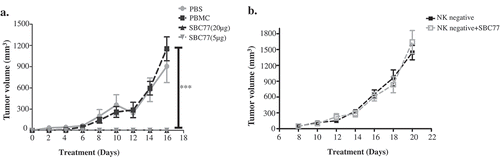
Figure 6. SBC77 inhibits tumor growth in vivo. (a) NOD/SCID mice (n = 5/group) were engrafted subcutaneously with LS174T cells (1 × 106 per animal) with freshly isolated human PBMCs (5 × 106 per animal). The mice then treated intraperitoneally with vehicle PBS (gray line), SBC77(20 µg/mice) (triangle, solid line), or SBC77 (5 µg/mice)(triangle, dashed line) every two days. The tumor volume was then measured. The data represent the average tumor volume of 5 mice. Error bars represent the standard deviation (***P < .001, t test, vehicle vs SBC77(20 µg, 5 µg)). (b) NOD/SCID mice (n = 5/group) were engrafted subcutaneously with LS174T cells (1 × 106 per animal) with freshly isolated human NK negative PBMC cells (5 × 106 per animal). The mice were then treated with SBC77 (20 µg/mice) or PBS as described in the Materials and Methods.