Figures & data
Figure 1. The functional role of FTX in OS. (a) QRT-PCR was used to examine the relative expression of FTX in OS tissues and ANT. (b) QRT-PCR result showed that FTX is up-regulated in all OS cell lines (U2OS, MG-63,143B, Saos-2), compared with normal cell line (HOS). (c) The transfected efficiency of shFTX#1 and shFTX#2 were examined by qRT-PCR. (d) Cell viability was assessed by CCK8 between NC FTX and shFTX in MG63 and 143B. (e) EdU was used to measure cell viability. (f) Colony formation was applied to explore cell viability. (g) Cell apoptosis was evaluated by TUNEL. (h) Flow cytometry was used to determine cell apoptosis. (i-j) Wound-healing assay and Transwell were used to explore cell migration ability. (k) Western blot was applied to measure protein expression level change. **P < .01.
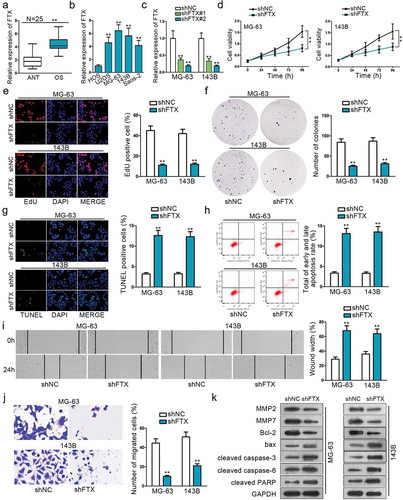
Figure 2. LncRNA FTX can interact with miR-320a. (a) MiR-320a was selected form 23 miRNAs that are combinative with FTX. (b) Subcellular fractionation assay and FISH indicated FTX was primarily located in cytoplasm. (c) Potential binding sites of FTX with miR-320a seed sequence on Starbase. (d) Dual-luciferase report proved the interaction between FTX and miR-320a. FTX WT referred to wild-type in FTX sequence to interact sequences of miR-320a pmirGLO luciferase reporter vector. FTX MUT was constructed using mutant binding sites. (e) RNA pull-down confirmed the physical interaction between FTX and miR-320a. (f-g) Relative expression of FTX in OS tissues and cells was observed by qRT-PCR. (h-j) CCK8, EdU and colony formation were used to inspect cell proliferation among shNC, shFTX and shFTX+ miR-320a inhibitor. (k-l) TUNEL and flow cytometry were used to examine cell apoptosis. (m-n) Wound healing and Transwell were used to measure cell migration. (o) Western blot was used to measure proteins expression level. *P < .05, **P < .01.
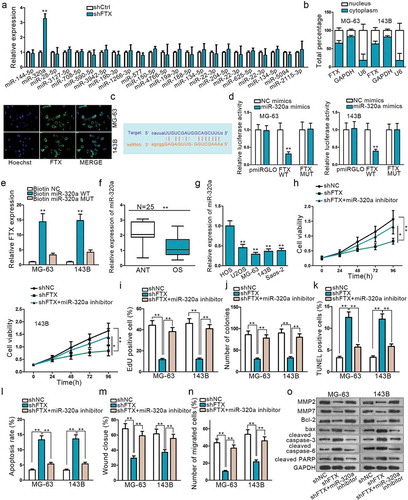
Figure 3. TXNRD1 is a potential target mRNA of miR-320a. (a) Use miR-320a mimics to select target gene TXNRD1. (b) The relative expression of TXNRD1 in OS tissues. (c) QRT-PCR was performed to determine the relative expression of TXNRD1 in one normal cell line (HOS) and four OS cell lines (U2OS, MG-63,143B, Saos-2). (d) Potential binding sides between miR-320a and TXNRD1 predicted on Starbase. (e-f) RIP and dual-luciferase report verified the interaction between miR-320a and TXNRD1. TXNRD1 WT was formed through sub-cloning separately the wild-type in TXNRD1 3ʹ-UTR sequence to interact sequences of miR-320a pmirGLO luciferase reporter vector. TXNRD1 MUT was constructed using mutant binding sites. (g) QRT-PCR and Western blot found were used to measure TXNRD1 mRNA and protein expression after miR-3320a overexpression. (h) shTXNRD1 transfection efficiency was examined by qRT-PCR. (i) CCK8 found shTXNRD1 transfection weakened cell viability. (j-k) TUNEL and flow cytometry were used to evaluate the effects of shTXNRD1 on apoptotic cells. (l-m) Wound-healing and Transwell were conducted to examine the effects of knockdown of TXNRD1 on cell migration. **P < .01.
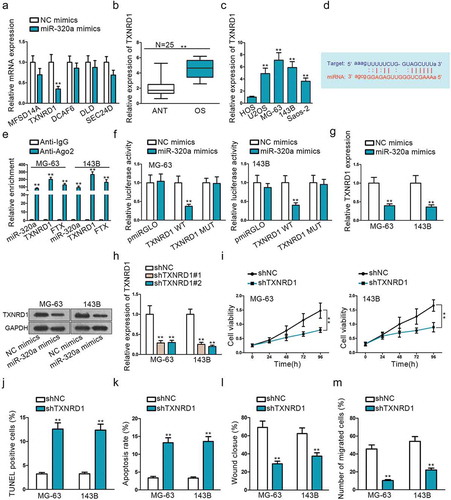
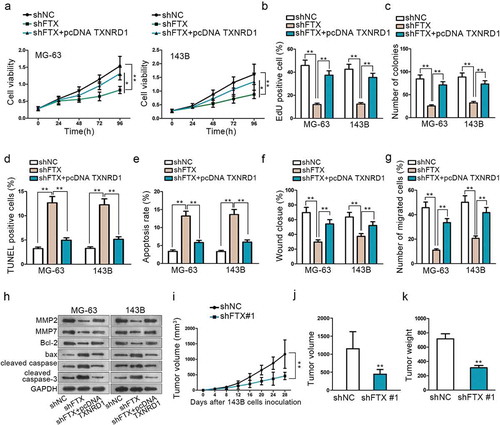
Figure 4. Knockdown of FTX can suppress OS development. (a-c) CCK8, EdU and colony formation assays were performed to detect cell proliferation among shNC, shFTX and shFTX+TXNRD1. (d-e) TUNEL and flow cytometry were used to examine OS cell apoptosis among shNC, shFTX and shFTX+TXNRD1. (f-g) Wound-healing and Transwell were performed to detect cell migration among shNC, shFTX and shFTX+TXNRD1. (h) Western blot was performed to measure protein expression level among shNC, shFTX, and shFTX+pcDNA TXNRD1. (i) Tumor growth curve in mice injected with shFTX than shNC. (j) Tumor volume in mice injected shNC and shFTX group at the end of inoculation. (k) Tumor weight injected shNC and shFTX group at the end of inoculation. *P < .05, **P < .01.