Figures & data
Table 1. Primers for real-time qPCR
Figure 1. The expression levels of FGF13 were significantly elevated in A549 cells, which might be benefit the cancer cells. (a) The localization of FGF13 was analyzed by confocal laser scanning microscope in A549 cells, and siFGF13 as negative controls. (b) Relative mRNA expression and (c) protein levels of FGF13 were detected in BEAS-2B and A549 cells, respectively. (d) The mRNA expression level and (e) protein levels of FGF13 were checked during A549 cells proliferation. Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01, *** P < .001
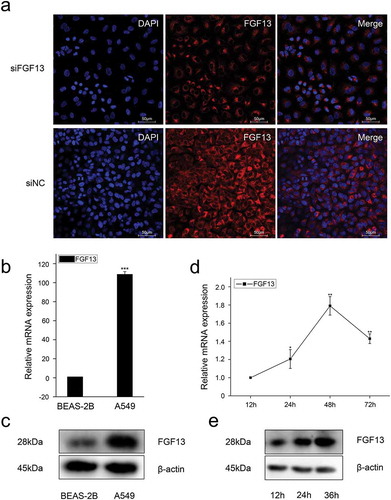
Figure 2. Knock-down of FGF13 inhibits A549 cells proliferation.A549 cells were transfected with siRNA-FGF13 or N.C siRNA, (a) the interference efficiency and relative mRNA expression levels of FGF13 were measured by real-time qPCR. (b) The protein expression level of FGF13 with the interference efficiency was detected by Western blotting. (c) Cell proliferation of A549 was measured by CCK8 assay at 0, 24, 48, 72 h post-transfection, respectively. (d) Cell colony formation was performed. (e) The number of A549 cells proliferation was checked by Ki67. (f) FGF13-silenced cells were harvested and samples were analyzed on the flow cytometer 72 h post-transfection. The cell numbers of G1 phase and S phase were counted in (f). Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
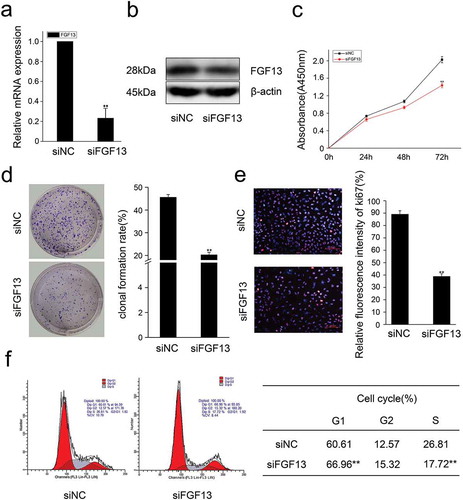
Figure 3. FGF13 accelerates cell cycle progression and promotes proliferation in A549 cells. A549 cells were transfected with pcDNA3.1-FGF13 (or Empty vector), (a) the over-expression efficiency and relative mRNA expression levels of FGF13 were measured by real-time qPCR. (b) the FGF13 relative protein levels of over-expression efficiency were detected by western blot after 48 h transfection. (c) Cell proliferation of A549 was measured by CCK8 assay at 0, 24, 48, 72 h post-transfection, respectively. (d) Cell colony formation was performed, (e) cell cycle progression was analyzed by the flow cytometer. The cell numbers of G1phase and S phase were counted in (e). Results were presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
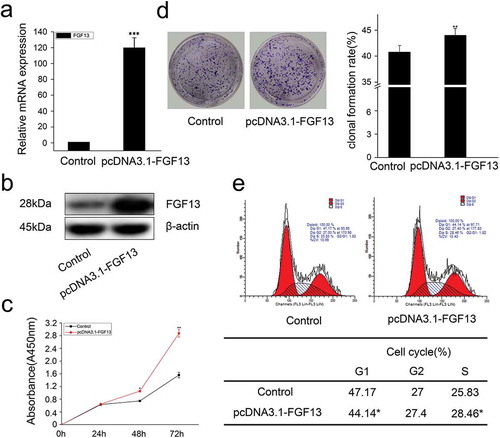
Table 2. Proteins that interacted with FGF13 in the yeast two-hybrid system
Figure 4. FGF13 shares a common distribution with SHCBP1 in the cytoplasm, which could bind to SHCBP1. (a) Cell localization of SHCBP1 was measured in A549 cells. (b) Colocalization of FGF13 and SHCBP1 was examined in A549 cells. (c) Co-IP assay was used to determine the interaction effects of FGF13 and SHCBP1. (d)The protein expression level of SHCBP1 with the interference efficiency was detected by Western blotting. (e)The CCK8 and (f) Ki67 assay were employed to assess A549 cells proliferation after SHCBP1 silencing. (g) The CCK8 assay was performed to assess the cell proliferation rate after co-interference 24 h, 48 h, 72 h. Results were presented as mean± S. E. M. n = 3. *P < .05, ** P < .01
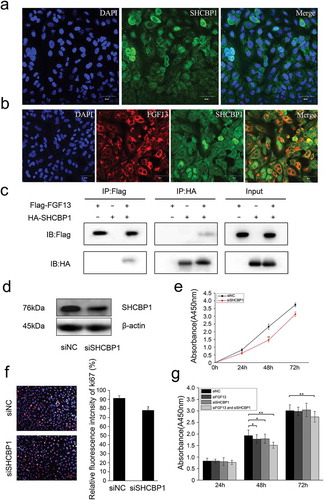
Figure 5. Interaction between FGF13 and SHCBP1 affects cell proliferation. FGF13 affects AKT signaling pathway, p21 and p27 expression levels were detected in A549 cells which overexpressed or interfered. (a) The mRNA expression levels of p21, p27 and CDK2 were analyzed on A549 cells with FGF13 silenced, the protein expression of p21, p27, cyclinE1 and FGF13 were detected with FGF13 silenced, respectively. (b) The mRNA expression levels of p21, p27 and CDK2 were evaluated in A549 cells with FGF13 overexpressed, the protein expression of p21, p27, cyclinE1 and FGF13 were detected with FGF13 overexpressed. (c) After FGF13 and SHCBP1 were silenced, the western blotting was employed to assess the expression of p21, p27 and cyclinE1, β-actin was used as a loading control. (d) FGF13 was overexpressed while SHCBP1 was interfered in A549 cells, the protein expression levels of p21, p27 and cyclinE1 were analyzed. The results of western blotting were assessed using ImageJ (mean ± SD of 3 independent experiments.*P < .05, ** P < .01)

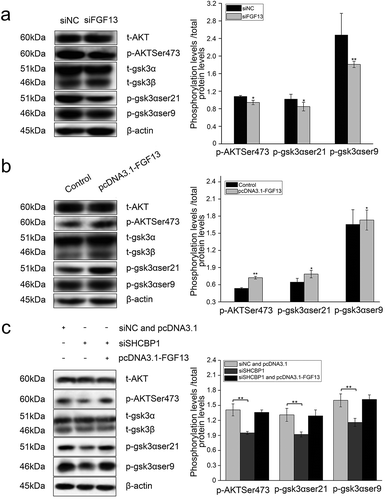
Figure 6. FGF13 activates AKT-GSK3 signaling pathway to regulate A549 cells proliferation. FGF13 and SHCBP1 co-interference or co-overexpression is performed in A549 cells, respectively. (a) After FGF13 were silenced, the protein levels of AKT, p-AKT (Ser473), GSK3α, p-GSK3α (ser21), GSK3β and p-GSK3β (ser9) were analyzed by Western blotting. (b) After FGF13 were overexpressed, AKT, p-AKT (Ser473), GSK3α,p-GSK3α(ser21), GSK3β and p-GSK3β(ser9) protein levels were detected in A549 cells.(c) FGF13 was overexpressed while SHCBP1 was interfered, the protein expression levels of AKT, p-AKT (Ser473), GSK3α, p-GSK3α (ser21), GSK3β and p-GSK3β (ser9) were analyzed., β-actin was used as a loading control. Results are presented as mean± S. E. M. n = 3. *P < .05, ** P < .01