Figures & data
Figure 1. MiR-199b-5p was oncogenic in MM. (a) GEO data analysis showed miR-199b-5p up-regulation in MM. (b) MiR-199b-5p expression in nPCs, RPMI-8266, NCI-H929, OPM-2 and U266 cells was estimated via qRT-PCR. (c) The transfection efficacy of miR-199b-5p inhibitor in OPM-2 and U266 cells was determined through qRT-PCR. (d-e) Cell proliferation of OPM-2 and U266 cells was tested by colony formation and EdU assays. (f-g) Cell apoptosis of OPM-2 and U266 cells was examined via TUNEL and flow cytometry analysis. (h) Western blotting of apoptosis-related proteins including Bcl-2, Bax, total and cleaved caspase-3 and caspase-9. *P < .05 and **P < .01 vs control group
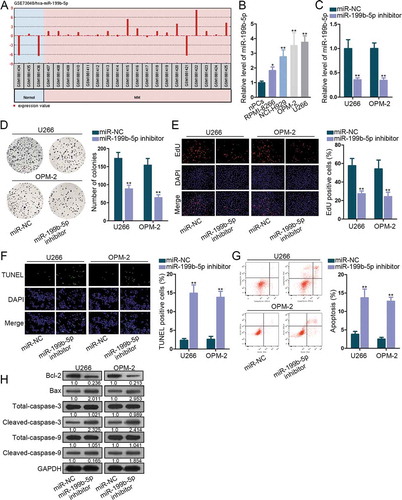
Figure 2. The interplay between LRRC75A-AS1 and miR-199b-5p. (a) Ten lncRNAs were screened out by starBase v3.0. (b) MS2-RIP screening of lncRNAs interacting with miR-199b-5p in OPM-2 and U266 cells. (c) qRT-PCR analysis of SNHG12, TMPO-AS1 and LRRC75A-AS1 levels in nPCs and MM cells. (d) FISH assay for the location of LRRC75A-AS1 in OPM-2 and U266 cells. (e) RNA pull-down analysis of miR-199b-5p interaction with LRRC75A-AS1. (f) The sequences of LRRC75A-AS1-WT/Mut and miR-199b-5p. (g) qRT-PCR analysis of miR-199b-5p overexpression in U266 and OPM-2 cells. (h) Luciferase reporter experiments were employed to validate the binding of miR-199b-5p to LRRC75A-AS1. (i) LRRC75A-AS1 was increased by pc-LRRC75A-AS1 as confirmed by qRT-PCR. (j-k) Colony formation and EdU assays examined the proliferation of OPM-2 and U266 cells. (l-n) TUNEL, flow cytometry and western blotting measured the apoptosis of OPM-2 and U266 cells. *P < .05 and **P < .01 vs control group
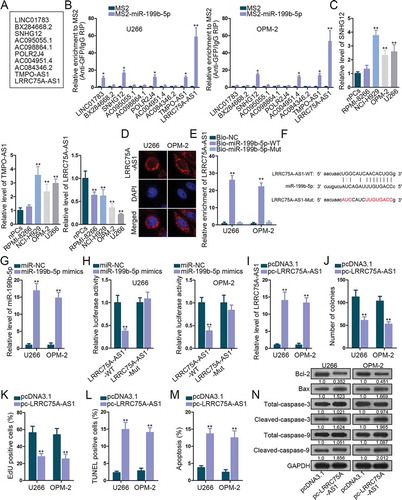
Figure 3. MiR-199b-5p targeted PDCD4 in MM cells. (a) The Venn diagram revealed the targets of miR-199b-5p identified by 4 prediction programs in Starbase. (b) Pie chart of the expression change rates of these genes in nPCs and MM cells. (c) Luciferase reporter assay in 293 T cells to test the binding affinity of six down-regulated genes in MM cells with miR-199b-5p. (d-e) qRT-PCR and western blotting detections of PDCD4 expression in MM cells in comparison with control cells or in MM cells transfected with miR-199b-5p mimics or miR-NC. (f) The binding between miR-199b-5p and LRRC75A-AS1 was testified by RNA pull-down. (g-h) Sequences of PDCD4-WT/Mut and miR-199b-3p. Luciferase reporter experiments was used for confirming the binding of miR-199b-3p at predicted sites in PDCD4. (i) The overexpression efficiency of pc-PDCD4 in U266 and OPM-2 cells was evaluated via qRT-PCR and western blotting. (j) Quantification of colonies formed by MM cells. (k) Quantification of EdU-labeled MM cells under PCDC4 overexpression. (l) Quantification of TUNEL-labeled MM cells upon PCDC4 overexpression. (m) Quantification of apoptotic MM cells in flow cytometry analysis. (n) Western blot of apoptosis-related genes in MM cells with PDCD4 overexpression. *P < .05 and **P < .01 vs control group
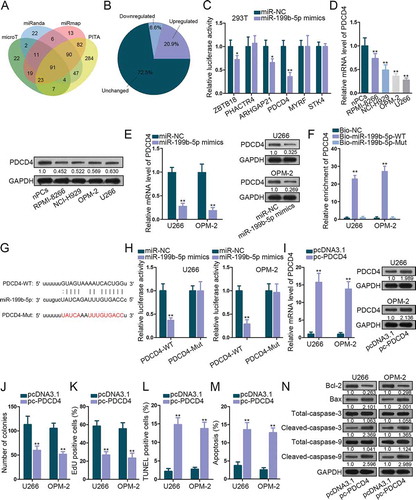
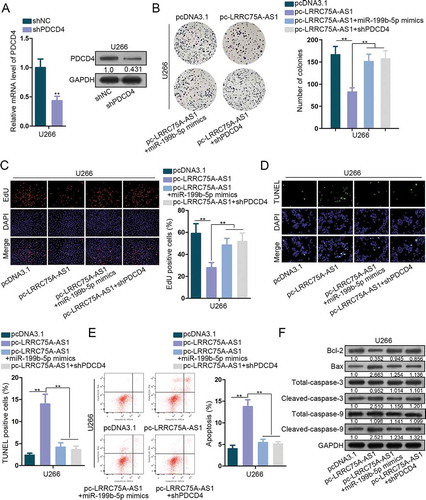
Figure 4. LRRC75A-AS1/miR-199b-5p/PDCD4 axis functioned in MM. (a) The inference efficiency of shPDCD4 in U266 cells for rescue assays was analyzed through both qRT-PCR and western blot. (b-c) MM cells were under transfection of pcDNA3.1, pc-LRRC75A-AS1, pc-LRRC75A-AS1 + miR-199b-5p mimics, or pc-LRRC75A-AS1 + shPDCD4. The proliferation of U266 cells in four groups was dissected via colony formation and EdU assays. (d-f) The apoptosis of U266 cells under four treatments was estimated via TUNEL, flow cytometry and western blot. **P < .01 vs control group