Figures & data
Figure 1. CYP2C8 was anti-proliferative and indicative of favorable survival in liver cancer. (a) The expression of CYP2C8 in LIHC tumor dataset (red) and normal dataset (gray) was plotted. (b) Kaplan-Meier survival analysis illustrated the relationship between CYP2C8 expression and overall survival from LIHC dataset. (c and d) LC cells from four cell lines and normal cells underwent qRT-PCR analysis to detect CYP2C8 mRNA (c) and protein (d) levels. (e) HepG2 and Huh-7 cells transfected with pcDNA3.1 or pcDNA3.1/CYP2C8 underwent qRT-PCR analysis to detect CYP2C8 mRNA level. (f) The proliferation of HepG2 and Huh-7 cells transfected with pcDNA3.1 or pcDNA3.1/CYP2C8 was tested by EdU assay. (g) CCK-8 assay monitored the reduced viability of HepG2 and Huh-7 cells after CYP2C8 overexpression. Optical density (OD) at 450 nm was regarded as the measurement of cell viability. (h) TUNEL assay evaluated the enhanced apoptosis of HepG2 and Huh-7 cells after the transfection of CYP2C8 overexpression plasmids. (i) Flow cytometry analysis examined the influence of CYP2C8 overexpression on LC cell apoptosis. p < .05 (*), p < .01 (**), p < .001 (***)
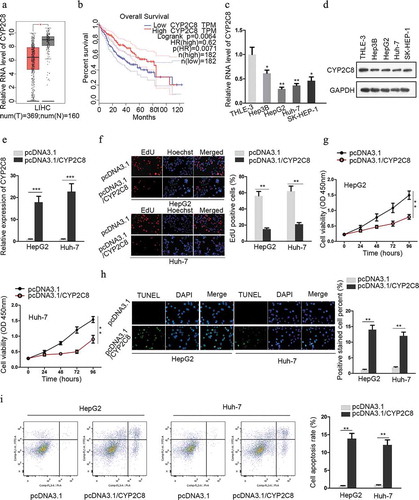
Figure 2. CYP2C8 was directly targeted by miR-382-3p. (a) According to the prediction of miRDB, top three miRNAs (miR-4797-5p, miR-382-3p and miR-3665) with higher target score with CYP2C8 were selected. The alteration of CYP2C8 mRNA levels in HepG2 and Huh-7 cells were evaluated in response to the transfection of mimics or inhibitors of these miRNAs. (b) CYP2C8 protein levels in LC cells were measured in response to miR-382-3p overexpression or inhibition by western blot. (c) RIP assay revealed that CYP2C8 mRNA and miR-382-3p could be enriched with Ago2 protein. (d) The sequences of wild-type CYP2C8 3ʹ-UTR, mutated CYP2C8 3ʹ-UTR and miR-382-3p were shown. The binding sites were highlighted in red. (e) Luciferase reporter assay showed that miR-382-3p overexpression inhibited the luciferase activity of CYP2C8-WT reporters in HepG2 and Huh-7 cells. (f) RNA pull-down demonstrated that CYP2C8 mRNA could be enriched by bio-miR-382-3p probes. p < .01 (**), p < .001 (***)
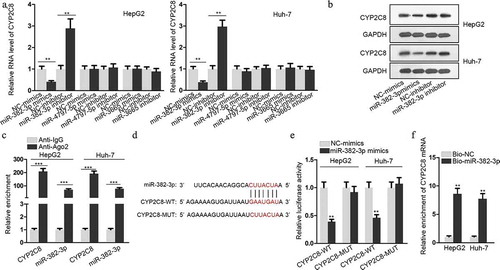
Figure 3. GAS5 bound to miR-382-3p to positively regulate CYP2C8 expression. (a) Top three lncRNAs (SNHG1, GAS5 and NORAD), which could possibly associate with miR-382-3p, were presented using ENCORI. Luciferase reporter assay evaluated the the luciferase activity of SNHG1-WT, GAS5-WT and NORAD-WT reporters after the transfection of NC-mimics or miR-382-3p mimics into HepG2 and Huh-7 cells. (b) FISH assay revealed GAS5 subcellular location in HepG2 and Huh-7 cells. (c) Cytoplasmic and nuclear RNA isolation uncovered the distribution of GAS5 in the cytoplasm or nuclei of HepG2 and Huh-7 cells. (d) lncLocator predicted a high possibility of the cytoplasmic abundance of GAS5. (e) RIP assay illustrated the enrichment of GAS5, miR-382-3p and CYP2C8 with Ago2 protein. (f) Complementary sequences between miR-382-3p and GAS5, together with the mutant site of GAS5 were shown. (g) Luciferase reporter assay showed the decline of GAS5-WT reporter luciferase activity due to the transfection of miR-382-3p mimics. (h) The impact of GAS5 overexpression on miR-382-3p level in LC cells was assessed by qRT-PCR. (i) qRT-PCR and western blot illustrated the effects of altered GAS5 expression on CYP2C8 mRNA and protein levels in LC cells. p < .01 (**), p < .001 (***), N.S.: not significant
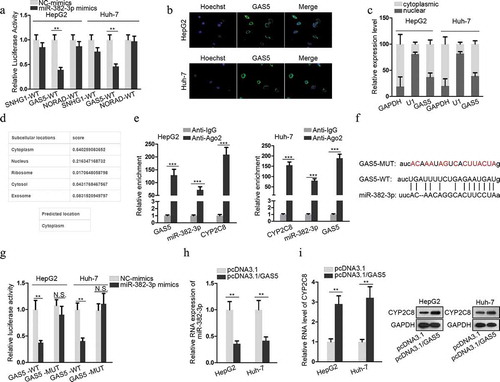
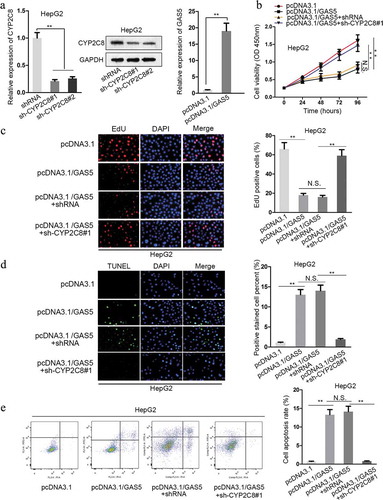
Figure 4. CYP2C8 elevation counteracted the impact of GAS5 on LC cell proliferation and apoptosis. (a) The knockdown efficiency of CYP2C8 and the overexpression efficiency of GAS5 in HepG2 cells were measured by qRT-PCR and western blot. (b) CCK-8 assay indicated that CYP2C8 knockdown reversed the inhibitory effect of GAS5 overexpression on HepG2 cell viability. (c) EdU assay showed that CYP2C8 silencing facilitated HepG2 cell proliferation which had been suppressed by GAS5 overexpression. (d) TUNEL assay presented that CYP2C8 knockdown abolished the enhanced HepG2 cell apoptosis altered by GAS5. (e) Flow cytometry analysis evaluated the cell apoptotic level of the transfected cells. p < .01 (**), N.S.: not significant