Figures & data
Table 1. Cell lines and culture medium used in this study
Table 2. qRT-PCR primer sequences
Figure 1. MiR-133b is lowly expressed in LUAD
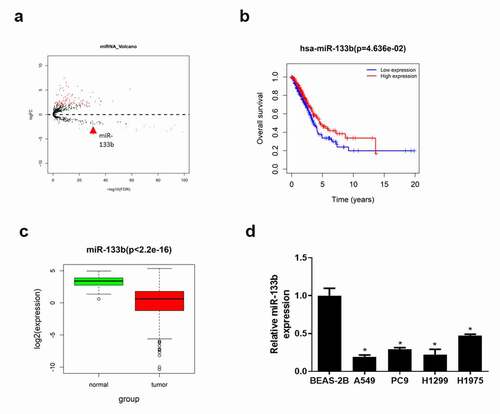
Figure 2. Overexpressing miR-133b suppresses the proliferation, migration and invasion of LUAD cells
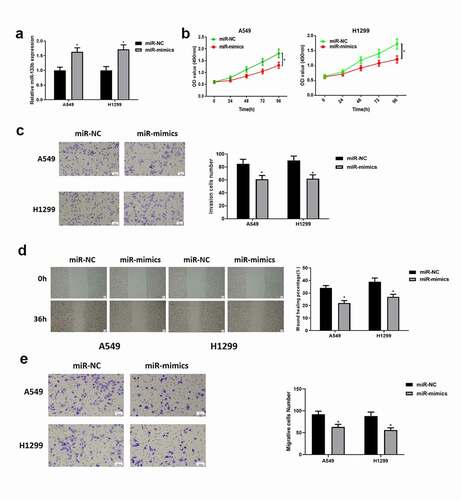
Figure 3. SKA3 is upregulated and correlated with poor prognosis in LUAD
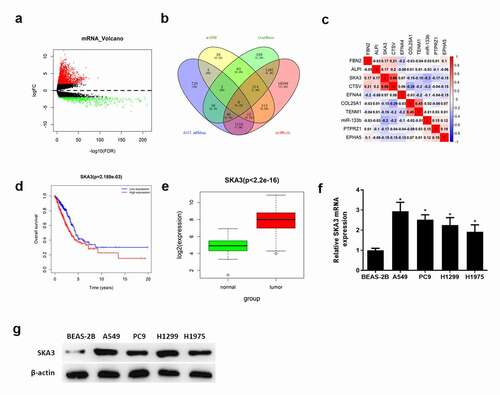
Figure 4. MiR-133b downregulates the expression of SKA3
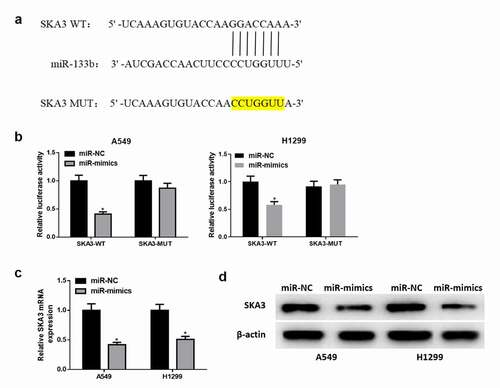
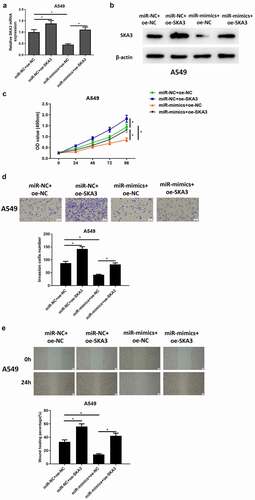
Figure 5. MiR-133b suppresses the proliferation, migration and invasion of LUAD cells through targeting SKA3