Figures & data
Figure 1. Knockdown of KDMs sensitizes resistant cells to CP. (a) and (c). CP-resistant A549 and 1703 cells were transfected with control siRNA (CsiRNA) or the indicated KDM siRNA and then treated with treated with vehicle (NT) or CP (1 µM) for 48 h. After drug removal, the cells were allowed to recover for 10 days. % colony formation is presented as graphs with SD indicated. There are significant differences (indicated with single asterisks above bars, p ˂0.05) between vehicle and CP treated conditions in some KDM siRNA transfected cells. There are significant differences in CP-treated A549 CPR cells between control siRNA and KDM3A siRNA (P = .002), KDM4B siRNA (P = .002), KDM5A siRNA (P = .018), and KDM6A siRNA (P = .000). There are significant differences in CP-treated 1703 CPR cells between control siRNA and KDM3A siRNA (P = .046), KDM4B siRNA (P = .05), KDM5A siRNA (P = .05), and KDM6A siRNA (P = .002) in A549. (b) and (d). qPCR analysis shows knockdown of the individual genes.
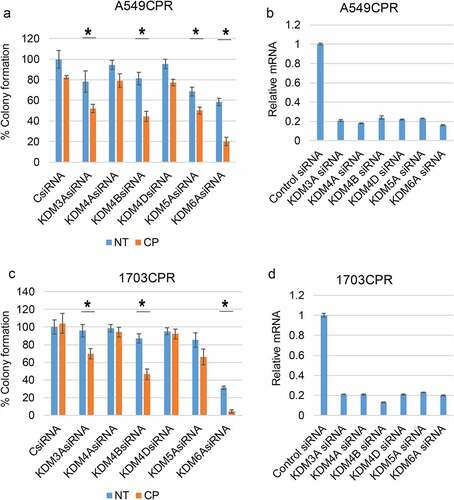
Figure 2. Inhibition of KDMs sensitizes resistant cells to CP and PTX. (a) Parental and resistant cells were treated with vehicle (NT) or CP (1 µM or 2 µM) with or without ML324 (10 µM) for 48 h. (b) Parental and resistant cells were treated with vehicle (NT) or CP (1 µM) with or without JIB04 (1 µM) for 48 h. Colony formation was determined 10d after drug removal. Triplicate results are shown. There are significant differences in CP treated conditions between parental and CPR cell lines (indicated with single asterisks above bars, p ˂0.05). There are significant differences between CP1 and CP1+ ML324 conditions in A549 (P = .000), ACPR (P = .000), 1703 (P = .000), 1703CPR (P = .012), 1975 (P = .033), and 1975CPR (P = .039) cells and between CP2 and CP2+ ML324 conditions in A549 (P = .035), ACPR (P = .000), 1703 (P = .021), 1703CPR (P = .000), 1975 (P = .045), 1975CPR (P = .027) cells. There are significant differences between JIB04 and CP1+ JIB04 conditions in A549 (P = .000), ACPR (P = .000), 1703 (P = .001), and 1703CPR (P = .002) cells. (c) A549 and A549CPR cells were treated with vehicle, CP (1 µM or 2 µM) with or without JIB04 (1 µM) for two days and analyzed for cell proliferation for 6 days with staining of cellular DNA. Average (quadruplicate) relative DNA is presented with SD indicated. There is a significant difference between CP1 treated A549 and ACPR cells (P = .038). There is a significant difference between CP1 and CP1+ JIB04 treated ACPR cells (P = .043). (d) Parental and resistant cells were treated with vehicle (NT) or PTX (5 nM or 10 nM)) with or without ML324 (10 µM) or JIB04 (1 µM) for 48 h. Colony formation was determined 10d after drug removal. Triplicate results are shown. There are significant differences between A549 and A549PTXR cells treated with PTX (indicated above bars). There are significant differences between PTX5 and PTX5+ ML324 in A549 (P = .041) and APTXR (P = .021) cells and between PTX10 and PTX10+ ML324 in A549 (P = .036) and APTXR (P = .028) cells. (e) A549 and A549PTXR cells were treated with vehicle, PTX (1 nM or 10 nM) with or without JIB04 (1 µM) for two days and analyzed for cell proliferation for 6 days with staining of cellular DNA. Average (quadruplicate) relative DNA is presented with SD indicated. There is a significant difference between PTX1 treated A549 and ACPR cells (P = .022). There is a significant difference between JIB04 and PTX1+ JIB04 treated A549 cells (P = .031) and ACPR cells (P = .046).
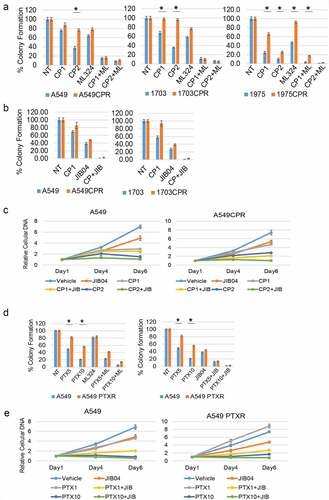
Figure 3. CP-resistant PDX tumors express higher KDMs. (a) Two mice bearing parental PDX tumors were treated with vehicle or CP (5 mg/kg) once and tumor growth was monitored. (b) PDX tumors were treated with five rounds of CP until the tumors became CP resistant. Two mice bearing CP-resistant PDX tumors were treated with vehicle or CP (5 mg/kg) once and tumor growth was monitored. (c) Expression of the indicated proteins in parental and CP-resistant PDX tumors were analyzed with immunoblot of tumor lysates.
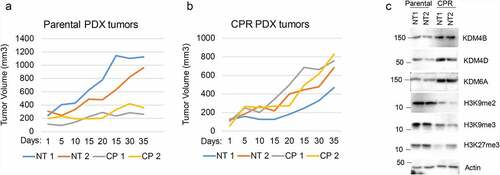
Figure 4. Combination JIB04 plus CP overcome CP-resistance in vivo. (a),(b) Mice bearing 1703 or 1703CPR tumors treated with vehicle (NT), CP (5 mg/kg, once), JIB04 (50 mg/kg/day, 5 days/week), or CP plus JIB04 and tumor growth monitored. There is a significant difference between vehicle and CP-treated 1703 tumors (P = .042). There is no significant difference between vehicle and CP (P = .25) and vehicle and JIB04 (P = .99) in 1703CPR tumors. There is a significant difference between vehicle and CP plus JIB04 (P = .003) treated 1703CPR tumors. There is a significant difference between CP plus JIB04 and CP (P = .043) or JIB04 (P = .049). (c) Tumor lysates (two tumors/group) were immunoblotted for the indicated proteins. (d) Average body weights of the mice bearing 1703CPR tumors are presented ± SE.
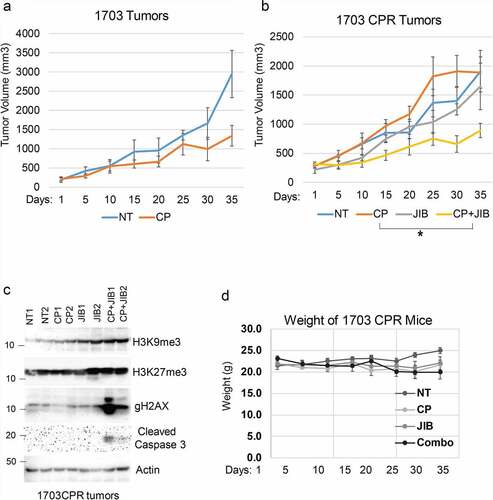
Figure 5. Jumonji KDM inhibitors reduce CtIP and PAF15 protein levels. 1703CPR and A549CPR cells were treated with CP (10 µM) and/or ML324 (10 µM), GSK-J4 (10 µM), or JIB04 (2 µM) for 24 hrs. Lysates were blotted for the indicated proteins.
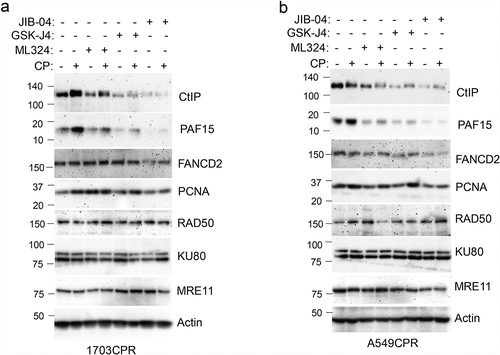
Figure 6. Knockdown of CtIP and PAF15 sensitizes cells to CP. (a) A549/A549CPR and 1703/a703CR cells were treated with vehicle or CP (10 µM) for 24 h. Lysates were immunoblotted for the indicated proteins. (b) 1703CPR cells were transfected with control siRNA (ctrl siRNA) or PAF15 or CtIP siRNA and then treated with vehicle or CP (10 µM) for 72 h. Apoptosis was assessed by determining the percentage of cells with sub-G1 DNA content by flow cytometry. Average (triplicate) % Sub-G1 cells (apoptosis) were presented with SD indicated. There are significant differences between control siRNA and PAF15 siRNA (P = .041) and between control siRNA and CtIP siRNA (P = .000) in CP treated conditions. (c) Lysates were immunoblotted for the indicated proteins. (d) 1703CPR cells were transfected with control siRNA or PAF15 siRNA and then treated with CP (5 µM) for 24 h. The cells were then grown in drug free media for another 48 h. Cells were harvested at the indicated time points and analyzed for cell cycle. Cell cycle histograms are presented.
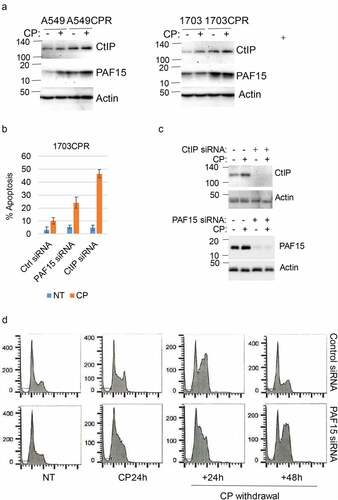
Figure 7. JIB04 reduces Rad51 Foci in CP-treated cells. 1703 CPR cells were treated with vehicle, CP, or CP plus JIB04 for 24 hrs. Cells were immunostained for Rad51. Rad51 foci were counted (100 cells/slide) and average (triplicate) % foci relative to the control are presented with SD indicated. There are significant differences between vehicle/CP and CP/CP+JIB04 conditions (P values indicated).
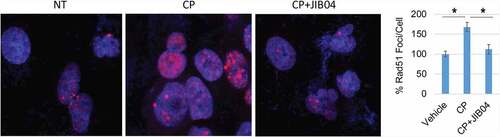
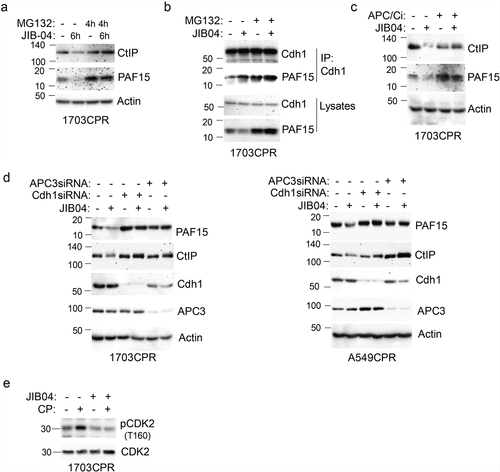
Figure 8. JIB04 induces APC/Cdh1-dependent degradation of CtIP and PAF15. a. 1703 CPR cells were treated with JIB04 (2 µM) for 6 h ± MG132 (10 µM) for 4 h and blotted. b. Lysates were IP’ed with Cdh1 antibodies and blotted for the indicated proteins. c. Cells were treated with JIB04 ± TAME (APC/Ci, 10 µM) for 6 h. d. Cells transfected with APC3 or Cdh1 siRNA were treated with JIB04 for 6 h and blotted. e. Cells were treated with CP and/or JIB04 for 24 h. Lysates were immunoblotted for the indicated proteins.