Figures & data
Figure 1. Vifs that are competent for APOBEC3H removal cannot induce cell cycle arrest. (A) Representative flow cytometry data showing VifNL4–3's or VifHXB2's potency in cell cycle regulation in HEK293T cells. (B) Bar chart representing the (G2/M)/G1 ratio of HEK293T cells transfected with VifNL4–3 or VifHXB2. Western blotting results (above) indicate the protein expression levels of VifNL4–3 and VifHXB2. (C) Western blotting results for APOBEC3H levels in the presence of VifHXB2 or VifNL4–3 in HEK293T cells. (D) Western blotting results for APOBEC3G levels in the presence of VifHXB2 or VifNL4–3 in HEK293T cells. (E) Western blotting results for APOBEC3F levels in the presence of VifHXB2 or VifNL4–3 in HEK293T cells. (F) Bar chart representing the (G2/M)/G1 ratio of HEK293T cells transfected with different Vifs that are potent in depleting APOBEC3H, with the exclusion of BNL43. (G) Western blotting results indicating the protein expression levels of the Vif proteins in (F).
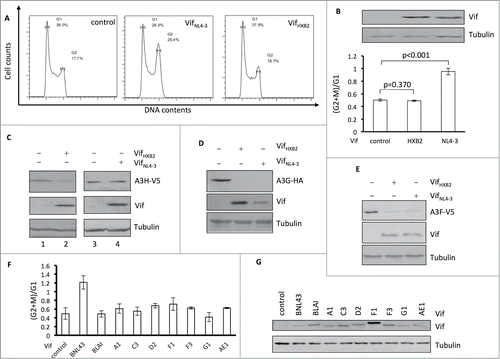
Figure 2. Vifs that are capable of G2 arrest cannot induce the depletion of APOBEC3H. (A) Representative flow cytometry data for cell cycle regulation by different Vifs in HEK293T cells. (B) Bar chart representing the (G2/M)/G1 ratios for HEK293T cells transfected with different Vif expression vectors. The Western blotting results (above) indicate the expression levels of Vif proteins. (C) Western blotting results for APOBEC3H levels in the presence of different Vif proteins in HEK293T cells. (D) Western blotting results for APOBEC3G levels in the presence of different Vif proteins in HEK293T cells
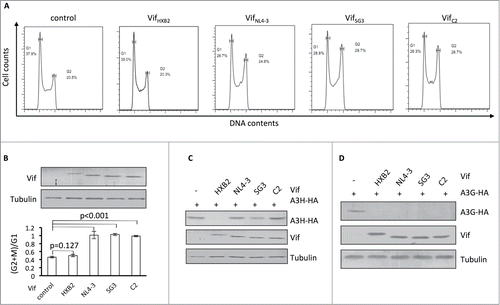
Figure 3. Position 48 is essential for Vif's induction of G2 arrest or depletion of APOBEC3H. (A) VifHXB2 and VifNL4–3 mutants used in this figure. (B) Bar chart representing the (G2/M)/G1 ratios in HEK293T cells transfected with VifHXB2, VifNL4–3, and their mutants at position 48. The Western blotting results (above) indicate the protein expression levels of VifNL4–3 and VifHXB2. (C) Bar chart representing the (G2/M)/G1 ratios in HEK293T cells transfected with VifHXB2, VifNL4–3, and their mutants at position 39. The Western blotting results (above) indicate the protein expression levels of VifNL4–3 and VifHXB2.
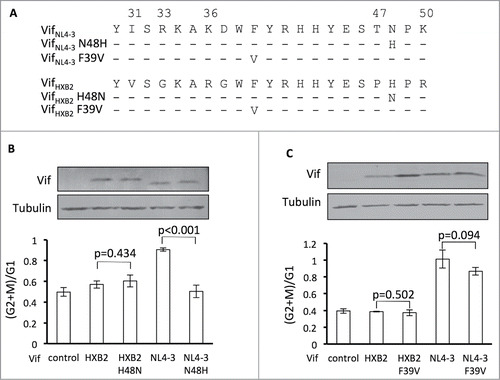
Figure 4. Positions 31, 33, 36, and 50, but not 47, are critical for Vif's induction of G2 arrest. (A) Protein alignment of G2-competent Vif proteins. G2-incompetent VifHXB2 was also introduced. The results suggest that position 47 is not conserved among different G2 arrest-competent Vifs. Numbers above the alignment indicate previously determined critical positions for Vif's induction of G2 arrest. Hyphens ("-") represent identical residues, as compared to VifNL4–3. (B) VifNL4–3 mutants used in this figure. (C) Representative flow cytometry data for cell cycle regulation by VifNL4–3 and its mutants. (D) Bar chart representing the (G2/M)/G1 ratios in HEK293T cells transfected with VifNL4–3 and its mutants. The Western blotting results (above) indicate protein expression levels of VifNL4–3.
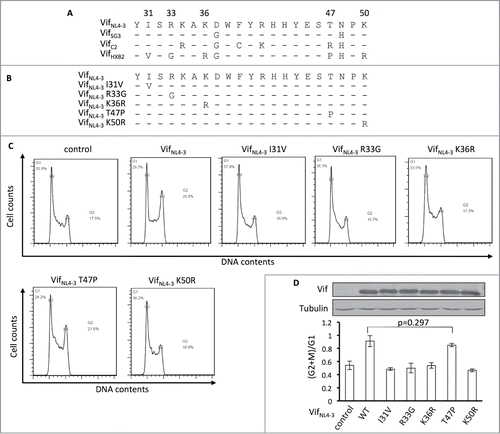
Figure 5. G2-related positions are also critical for Vif's induction of APOBEC3H depletion. (A) VifHXB2 and VifNL4–3 mutants used in this figure. (B) Western blotting results for APOBEC3H levels in the presence of VifHXB2 and its mutants at G2-related positions. VifNL4–3 was introduced as a negative control. (C) Western blotting results for APOBEC3H levels in the presence of VifHXB2, VifNL4–3, and their M5 mutants (with positions 31, 33, 36, 47, and 50 exchanged between VifHXB2 and VifNL4–3).
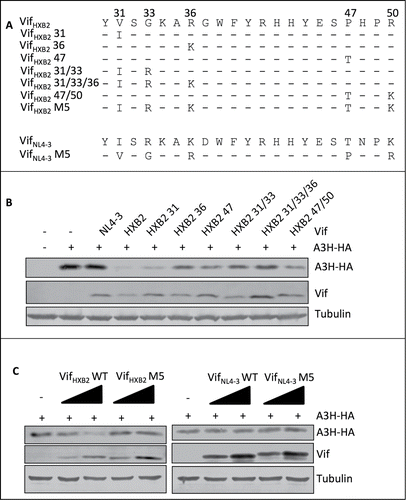
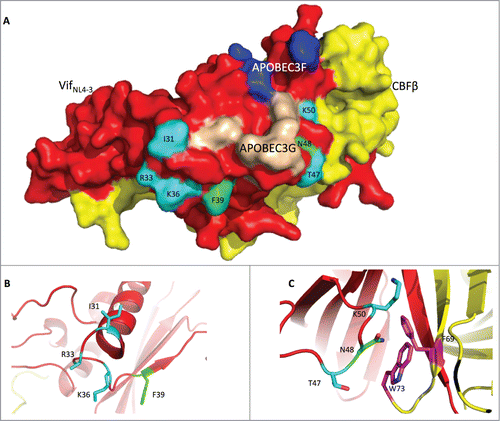
Figure 6. The Vif protein uses similar regions to induce G2 arrest and the degradation of APOBEC3H. (A) Surface model of the Vif-CBFβ interaction based on the published structure (PDB: 4N9F).Citation19 The Vif protein is labeled in red, and CBFβ is in yellow. On the Vif protein, the APOBEC3F-binding region is labeled in blue, the APOBEC3G-binding region in wheat, the critical residues for G2 arrest in cyan, and the critical residues for APBOEC3H depletion in green. (B) Detailed conformation of Vif around positions 31, 33, 36 (essential for G2 arrest), and 39 (essential for APOBEC3H depletion). (C) Detailed conformation of Vif around positions 47, 50 (for G2 arrest), and 48 (for APBOEC3H depletion). It is noteworthy that N48 on Vif may interact with F69 and W73 (in purple) on CBFβ.