Figures & data
Figure 1. Overexpression and amplification of PTP1B in gastric cancer. (A) PTP1B mRNA expression was significantly up-regulated in primary gastric cancers (T) as compared with matched normal gastric tissues (N) as determined by qRT-PCR assay (n = 29). PTP1B expression level was normalized with 18S rRNA level. Horizontal lines indicate mean ± SE. **, P < 0.01. (B) Real-time quantitative PCR was performed to analyze PTP1B copy number in a cohort of gastric cancers (T) and control subjects (N). Horizontal lines indicate mean ± SE. ***, P < 0.001. (C) Increasing extent of specific staining (brown color) was associated with increasing PTP1B copy number (number inside brackets). Shown are representative cases of immunohistostainging on gastric cancer histologic slides using anti-PTP1B antibody (upper panel). Linear regression analysis was performed to assess the relationship between immunohistostaining score and PTP1B copies on 12 randomly selected gastric cancer cases (lower panel, R = 0.73).
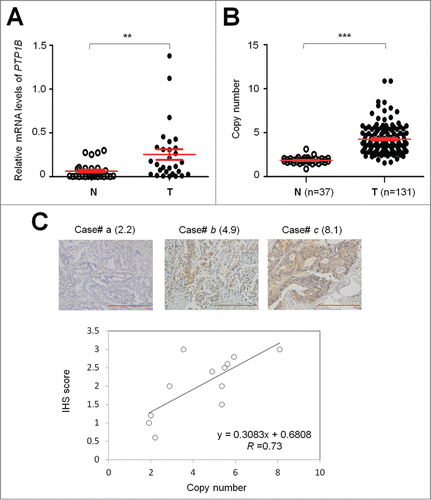
Table 1. Association of PTP1B amplification with clinicopathologic variables in gastric cancer
Table 2. PTP1B amplification in gastric cancer―multivariable models assessing age, differentiation, tumor invasion, tumor stage, lymph node metastasis and survival status
Table 3. Prognostic value of clinicopathologic factors and PTP1B amplification in univariate and multivariate Cox regression analysis (HR† and 95% CI)
Figure 2. Association of PTP1B amplification with poor survival of gastric cancer patients. Kaplan–Meier survival curves were used to assess the survival of primary gastric cancer patients. (A) The patients with PTP1B amplification (n = 68) had significantly shorter survival times than the patients without PTP1B amplification (n = 63). Median survival was 55.9 months among the patients with PTP1B amplification compared with 84.8 months among the patients without PTP1B amplification. The hazard ratio was 2.1 (95% confidence interval = 1.3, 3.5; P = 0.003, log rank test). (B) When the patients with residual cancers were excluded, the patients with PTP1B amplification (n = 57) still had significantly poor survival compared with the patients without PTP1B amplification (n = 60). Median survival was 59.8 months in the former compared with 88.1 months in the latter. The hazard ratio was 2.2 (95% confidence interval = 1.2, 3.7; P = 0.005, log rank test). Am+, PTP1B amplification; Am-, no amplification.
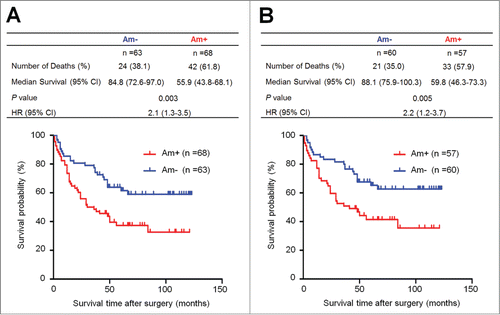
Figure 3 (See previous page). Inhibition of cell growth and induction of cell cycle arrest and apoptosis by knocking down PTP1B expression in gastric cancer cells. (A) Knockdown of PTP1B mRNA (upper panel) and protein (lower panel) by using 2 different siRNAs (si-PTP1B #1 and #2) in gastric cancer cell lines SGC7901 and MGC803 was evidenced by qRT-PCR and western blot, respectively. 18S rRNA was used as a normalized control for qRT-PCR assay. GAPDH was used as loading control in western blot analysis. ***, P < 0.001. (B) PTP1B downregulation significantly inhibited cell proliferation in gastric cancer cells. **, P < 0.01; ***, P < 0.001. (C) The effect of PTP1B knockdown on cell growth was further confirmed by colony formation assay. Left panel shows the representative images of colony formation in cells transfected with si-PTP1B #1 or si-NC. Quantitative analysis of colony numbers is shown in right panel. Data were presented as mean ± SE of values from 3 different assays. **, P < 0.01. (D) Cells were transiently transfected with si-PTP1B #1 or si-NC. After 72 h post-transfection, DNA content was measured by flow cytometry to determine cell cycle fractions. Representative flow cytometric histograms of cell transfected with si-PTP1B #1 and si-NC were shown in left panel. The fraction of cells in each cell cycle phase was indicated in right panel. Data were presented as mean ± SE of values from 3 independent experiments. *, P < 0.05; **, P < 0.01. (E) Cells transiently transfected with si-PTP1B #1 or si-NC. Cell apoptosis was then measured 72 h after transfection by flow cytometry analysis of Annexin V-FITC/PI double-labeled cells. Flow cytometry profiles represent SGC7901 and MGC803 cells with Annexin V-FITC staining in x axis and Pl in y axis, respectively (left panel). The percentages of early apoptotic (bottom right quarter) and late apoptotic (top right) cells were presented in the figures. The percentage of apoptotic cells was presented in right panel. The experiment was repeated 3 times and data were presented as mean ± SE. ***, P < 0.001.
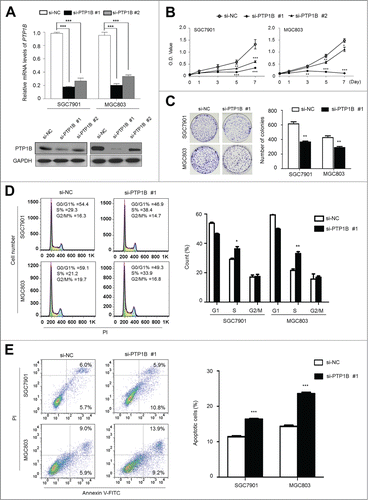
Figure 4. Inhibition of gastric cancer cell migration and invasion by down-regulating PTP1B expression. (A) Cells transfected with si-PTP1B #1 or si-NC were starved overnight and then seeded in the Transwell chambers without Matrigel for migration assay, and coated with Matrigel for invasion assay, respectively. After a 12 h or 24 h-culture, non-migrating (or non-invading) cells in the upper chamber were removed and migrating (or invading) cells were stained and calculated in 5 microscopic fields per sample. Shown are representative images of migrating (or invading) cells (left panels). The bar graphs (right panels), corresponding to left panels, show means ± SE of the numbers of migrating (or invading) cells from 3 independent assays. **, P < 0.01; ***, P < 0.001. (B) qRT-PCR assay was performed to assess the effect of PTP1B knockdown on the expression of metastasis-related genes MMP-2,-9 and -14 in gastric cancer cells. Expression levels of these genes were normalized with 18S rRNA levels. Data were presented as mean ± SE. *, P < 0.05;**, P < 0.01.
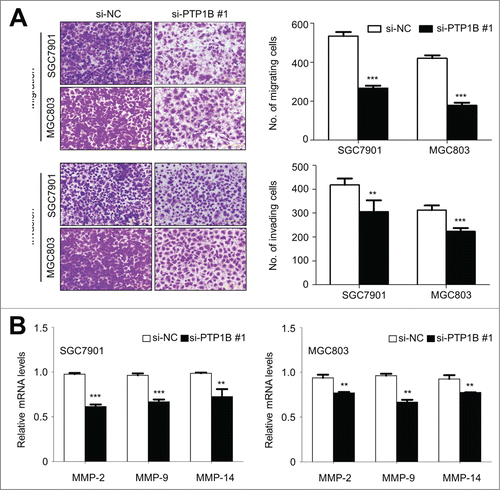
Figure 5 (See previous page). Effect of PTP1B down-regulation on the process of EMT in gastric cancer cells. Cells transfected with si-PTP1B #1 or si-NC were seeded on the coverslips in 6-well plates. After a 24 h-culture, immunofluorescence staining was performed to assess the expression of an epithelial cell marker E-cadherin and 2 mesenchymal markers N-cadherin and Vimentin in SGC7901 and MGC803 cells. Representative immunofluorescence stainings of target protein in cells transfected with si-PTP1B #1 and si-NC were shown in panel (A). Red color represents target protein fluorescence and blue color represents Hoechst33342 staining for nuclei. The percentage of cells showing positive E-cadherin, N-cadherin and Vimentin expression was indicated in panel (B). The experiment was repeated 3 times and data were presented as mean ± SE. ***, P < 0.001.
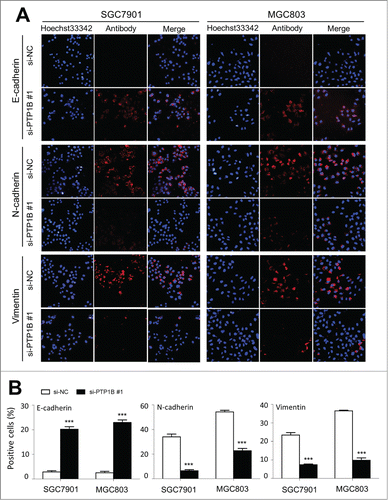
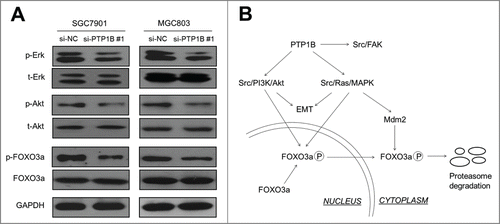
Figure 6. PTP1B acts as a potential oncogene by activating major signaling pathways. (A) Cells transfected with si-PTP1B #1 or si-NC were lysed and lysates were subjected to protein gel blot assays. The antibodies against phospho-Erk (p-Erk), total Erk (t-Erk), phospho-Akt (p-Akt) and total Akt (t-Akt) were used to determine the effect of PTP1B downregulation on the activities of the MAPK and PI3K/Akt pathways. The antibodies against FOXO3a and phosphorylated FOXO3a (p-FOXO3a) were used to test the effect of knocking down PTP1B expression on the transcriptional activity of FOXO3a. GAPDH was used as a loading control. (B) Schematic model of molecular mechanisms underlying oncogenic role of PTP1B in gastric cancer. PTP1B overexpression or amplification in gastric cancer cells increases Src activity by reducing phosphorylation at tyrosine 530 of Src and sequentially activates Src-related signaling pathways, such as Src/FAK, Src/Ras/MAPK and Src/PI3K/Akt. Moreover, knocking down PTP1B expression inhibits phosphorylation of FOXO3a, nuclear-cytoplasmic translocation and proteasome degradation through blockade of the MAPK and PI3K/Akt pathways, further contributing to the enhancement of FOXO3a transcriptional activity in gastric cancer cells. Taken together, PTP1B overexpression or amplification promotes gastric cancer cell growth and invasiveness through activating Src-related signaling pathways, ultimately contributing to poor prognosis of gastric cancer patients.