Figures & data
Figure 1. Assembly of pre-RC onto a plasmid DNA, depending on GAL4 DNA-binding sites. (A) Schematic of pre-RC assembly onto a plasmid DNA, depending on the binding sites. Plasmids attached to magnetic beads were used for the assembly of pre-RC. (B) Binding of GAL4 Cdc6, Mcm2, and Orc2 to the plasmids. Plasmids with various copy numbers of the GAL4 DNA-binding site (6×, 1×, 0×) were attached to the magnetic beads and incubated in a binding buffer with GAL4-Cdc6 on ice for 10 min. After incubation, the bead-coupled plasmids were isolated, washed with a high-salt buffer containing 350 mM NaCl and 100 mM KCl, and incubated in Cdc6-depleted egg extracts (ΔCdc6 LSS) at 23°C for 30 min. The plasmids were then isolated and washed with a low-salt buffer containing 100 mM KCl. Proteins bound to the plasmids were separated by SDS-PAGE and analyzed by immunoblotting. (C) Mcm2 binding to the plasmids, depending on the bound GAL4-Cdc6. The bead-coupled plasmids with or without the DNA-binding sites were pre-incubated in a binding buffer containing GAL4-Cdc6 or GAL4-GST on ice for 10 min. The washed plasmids were then incubated in ΔCdc6 LSS at 23°C for 30 min to assemble pre-RC on the plasmid. Proteins bound to the plasmids were analyzed. (D) Inhibition of Mcm2 binding to the plasmids by geminin. The bead-coupled plasmids bound with GAL4-Cdc6 were incubated in ΔCdc6 LSS in the presence or absence of 1 μM GST-geminin. The plasmids without GAL4-binding sites were used as a negative control (lane 1). (E) ORC-dependent Mcm2 binding to the plasmids bound with GAL4-Cdc6. The bead-coupled plasmids bound with GAL4-Cdc6 were incubated in Cdc6-depleted (ΔCdc6) or Cdc6 and Orc2 double-depleted (ΔOrc2ΔCdc6) LSS at 23°C for 30 min. The proteins in the extracts (extract) and bound to the plasmids (plasmid) were analyzed. (F) High-salt-wash-resistant binding of Mcm2 to the plasmids bound with GAL4-Cdc6. The bead-coupled plasmids incubated in ΔCdc6 LSS were isolated and washed with either the low-salt (100 mM KCl; L) or the high-salt (250 mM KCl; H) buffer after the GAL4 pre-RC was assembled on the plasmids. Proteins bound to the plasmids were analyzed.
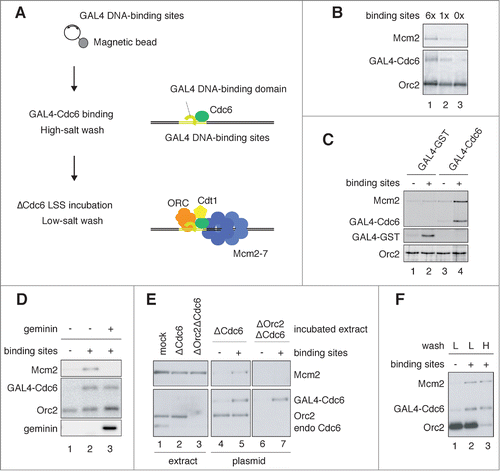
Figure 2. Assembly of GAL4 pre-RC on the plasmids proximal to GAL4 DNA-binding sites. (A) Schematic diagram of the 7.6 kbp plasmid. The location of 4 primer sets (X, Y, Y′, and Z) used for the ChIP analyses, and 6× GAL4 DNA-binding sites (light green) are indicated. (B) ChIP analysis of pre-RC components bound to the plasmids. The bead-coupled plasmids with (+) and without (-) the DNA-binding sites were incubated with GAL4-Cdc6, and the pre-RC was assembled as described in . The localization of pre-RC components was examined by ChIP. Antibodies used for immunoprecipitation are indicated at the top of each panel. The amount of target DNA in each IP sample was determined by qPCR, and relative enrichment was estimated by dividing the amount obtained with each IP sample by that of the mock IP sample using control antibody. Average data are shown as a bar graph with standard deviation (SD) indicated by bars from 3 independent PCR reactions of each ChIP sample. (C) Effect of geminin on the assembly of GAL4 pre-RC on the plasmids. The bead-coupled plasmids with the DNA-binding sites were pre-incubated with GAL4-GST (GST) or GAL4-Cdc6 (Cdc6), and the assembly of pre-RC components in the presence (+) and absence (−) of GST-geminin (1 μM) was analyzed as in (B).
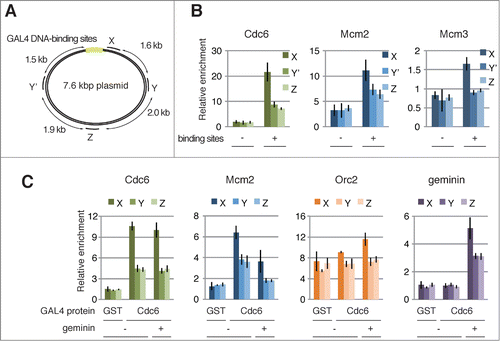
Figure 3. Replication of the plasmid DNA assembled with pre-RCs in NPE. (A) DNA replication products analyzed by agarose gel electrophoresis. Conventional pre-RC and GAL4 pre-RC were assembled by incubating the bead-coupled plasmids (3.0 kbp) at 23°C for 30 min in mock-depleted and Cdc6-depleted HSS, respectively. We then added 2 vol of NPE containing [α32P]-dATP to 1 vol of the assembly mixture, and the reaction was terminated at the indicated time by isolating DNA. The purified DNA was separated by agarose gel electrophoresis, and the replication products were visualized by autoradiography. The plasmid without the GAL4-binding sites was used as a negative control (-preRC) and 500 μM roscovitine was added to inhibit the initiation of DNA replication (+). (B) Graphic presentation of quantified data from (A). Total amounts of [α32P]-dATP incorporated into DNA were measured and plotted against incubation time in NPE.
![Figure 3. Replication of the plasmid DNA assembled with pre-RCs in NPE. (A) DNA replication products analyzed by agarose gel electrophoresis. Conventional pre-RC and GAL4 pre-RC were assembled by incubating the bead-coupled plasmids (3.0 kbp) at 23°C for 30 min in mock-depleted and Cdc6-depleted HSS, respectively. We then added 2 vol of NPE containing [α32P]-dATP to 1 vol of the assembly mixture, and the reaction was terminated at the indicated time by isolating DNA. The purified DNA was separated by agarose gel electrophoresis, and the replication products were visualized by autoradiography. The plasmid without the GAL4-binding sites was used as a negative control (-preRC) and 500 μM roscovitine was added to inhibit the initiation of DNA replication (+). (B) Graphic presentation of quantified data from (A). Total amounts of [α32P]-dATP incorporated into DNA were measured and plotted against incubation time in NPE.](/cms/asset/f874d33e-ee93-4a3d-9560-df14befe99b6/kccy_a_1007003_f0003_b.gif)
Figure 4. Site-specific assembly of the pre-IC and replisome components and initiation of DNA replication. (A) Recruitment of initiator proteins onto the bead-coupled plasmids assembled with pre-RCs. The plasmids assembled with the conventional or the GAL4 pre-RC were incubated in NPE supplemented with 1 ng/μl aphidicolin at 23°C for 30 min, and 500 μM roscovitine was used for inhibiting the initiation of DNA replication (+ roscovitine). The plasmids bound with GAL4-GST were used as a negative control of GAL4 pre-RC assembly (-pre-RC). Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) Localization of the proteins bound to the plasmids proximal and distal to the DNA-binding sites. The bead-coupled plasmids bound with GAL4-GST (-pre-RC) or GAL4-Cdc6 (GAL4 pre-RC) were incubated in Cdc6-depleted HSS at 23°C for 30 min and further incubated for 30 min after addition of NPE containing 1 ng/μl aphidicolin. We added 1 μM p21 to inhibit the initiation of DNA replication (p21 +). ChIP analysis was performed using antibodies as indicated at the top of the panels. The amount of target DNA in each IP sample was determined by qPCR using primer sets at the proximal (X) and distal site (Z), and relative enrichment was estimated as described in . (C) Identification of the initiation sites by BrdUTP incorporation into the plasmids assembled with GAL4 pre-RC. The plasmids with or without pre-RC (GAL4 pre-RC or -pre-RC, respectively) were incubated in NPE supplemented with 1 ng/μl aphidicolin and 200 μM BrdUTP at 23°C for 15 min. Roscovitine (500 μM) was added to inhibit the initiation of DNA replication (roscovitine +). The incorporation reaction was terminated by isolating DNA and the isolated DNA was then fragmented, followed by immunoprecipitation with anti-BrdU antibody. Enrichment of DNA fragments incorporated with BrdUTP was determined by qPCR with 3 primer sets (X, Y, and Z). The amounts of target DNA in each IP sample relative to the mock IP sample, both of which were determined by qPCR, were taken as relative enrichment. Average data are shown as bar graphs with standard deviation (SD) indicated by bars from 3 independent PCR reactions of each IP sample.
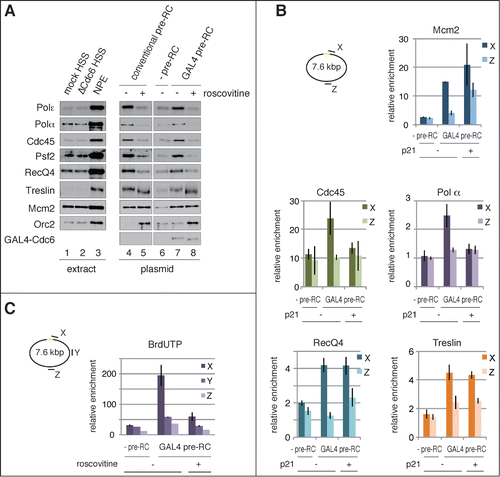
Figure 5. Unwinding of a site-specific origin upon activation of GAL4 pre-RC. (A) Schematic of P1 nuclease assay for unwinding of plasmid DNA after activation of pre-RCs in NPE. P1-treated DNA was further digested with BamHI or MfeI that recognizes different unique digestion sites of the plasmid. BamHI and MfeI sites are located at the proximal side (at the edge; 0 kb distance) and opposite side (3.0 or 3.3 kb distance) of the DNA-binding sites, respectively. (B) The bead-coupled plasmids assembled with conventional (C) or GAL4 pre-RC (G) were incubated in NPE containing 67 ng/μl aphidicolin at 23°C for 30 min to allow unwinding of dsDNA. Plasmids bound with GAL4-GST were used as a negative control of pre-RC assembly (N). After incubation, the plasmids were isolated and treated with P1 nuclease to digest ssDNA regions of the plasmids. P1-treated DNA was then purified and further digested with BamHI or MfeI. The purified DNA from the restriction enzyme digestions was subjected to agarose gel electrophoresis and visualized. (C) The distribution of DNA fragments obtained after P1 nuclease followed by the restriction enzyme digestions from a single experiment. The amount of DNA was estimated from the staining of DNA in agarose gel with the aid of the Image J program and shown as brightness. The peak values of each sample were taken as 100%. Reproducibility of the data is shown in Figure S5.
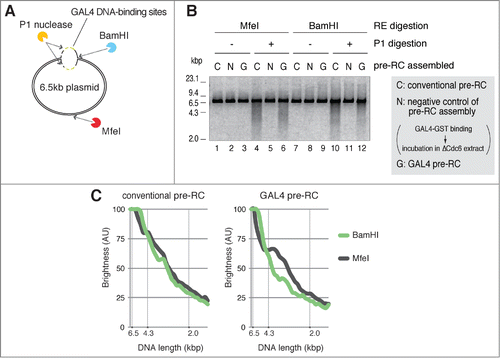
Figure 6 (See previous page). RecQ4-dependent assembly of replisome components, initiation of replication, and unwinding of dsDNA. (A) RecQ4-dependent and -independent binding of initiator proteins to the plasmids. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted HSS at 23°C for 30 min to allow pre-RC assembly. Subsequently, 2 vol of mock-depleted NPE with or without 1 μM p21, or RecQ4-depleted NPE with or without 530 nM N-terminal fragment of RecQ4 (N2), was added to 1 vol of each corresponding HSS and further incubated for 30 min. Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) RecQ4 promotes replication of the plasmids in NPE. The bead-coupled plasmids (3.0 kbp) assembled with pre-RC were incubated in NPE as shown in , except for the presence of [α32P]-dATP. The reactions were terminated at the indicated time and the incorporation of [α32P]-dATP into DNA was detected after separating DNA by agarose gel electrophoresis. (C) RecQ4-dependent and -independent recruitment of replication proteins to the site-specific origin. The bead-coupled plasmids (7.6 kbp) bound with GAL4-Cdc6 were incubated at 23°C for 30 min in Cdc6-depleted or Cdc6 and RecQ4 double-depleted HSS. After the incubation, mock-depleted or RecQ4-depleted NPE containing 1 ng/μl aphidicolin was added to the corresponding HSS and further incubated for 30 min. The localization of proteins bound to the plasmids was examined by ChIP analysis as described in , except for using primer sets at X and Z, proximal and distal to the origin, respectively. Antibodies used for ChIP are indicated at the top of each panel. (D) Stable binding of Cdc45, Mcm2-7, and GINS to the plasmid both in the presence and absence of RecQ4. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted extracts as described in . After incubation in the mock- or RecQ4-depleted NPE, plasmid-beads were isolated and washed with the buffer containing various concentrations of NaCl. Proteins bound to the plasmids (plasmid) and in the extracts (extract) were analyzed by immunoblotting. (E) RecQ4 promotes unwinding of DNA in NPE. The bead-coupled plasmids (7.6 kbp) were incubated at 23°C for 30 min in RecQ4-depleted HSS, and then 2 vol of RecQ4-depleted NPE was added with and without a 720 nM N-terminal fragment of RecQ4 (N2) in the presence of 67 ng/μl aphidicolin. After 30 min incubation, the plasmids were isolated and treated with P1 nuclease at 38°C for 10 min. Total DNA isolated from the mixture was further digested with BamHI, and the purified DNA was separated by agarose gel electrophoresis. As a positive control, mock-depleted extracts were used for the unwinding assay (mock).
![Figure 6 (See previous page). RecQ4-dependent assembly of replisome components, initiation of replication, and unwinding of dsDNA. (A) RecQ4-dependent and -independent binding of initiator proteins to the plasmids. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted HSS at 23°C for 30 min to allow pre-RC assembly. Subsequently, 2 vol of mock-depleted NPE with or without 1 μM p21, or RecQ4-depleted NPE with or without 530 nM N-terminal fragment of RecQ4 (N2), was added to 1 vol of each corresponding HSS and further incubated for 30 min. Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) RecQ4 promotes replication of the plasmids in NPE. The bead-coupled plasmids (3.0 kbp) assembled with pre-RC were incubated in NPE as shown in Figure 6A, except for the presence of [α32P]-dATP. The reactions were terminated at the indicated time and the incorporation of [α32P]-dATP into DNA was detected after separating DNA by agarose gel electrophoresis. (C) RecQ4-dependent and -independent recruitment of replication proteins to the site-specific origin. The bead-coupled plasmids (7.6 kbp) bound with GAL4-Cdc6 were incubated at 23°C for 30 min in Cdc6-depleted or Cdc6 and RecQ4 double-depleted HSS. After the incubation, mock-depleted or RecQ4-depleted NPE containing 1 ng/μl aphidicolin was added to the corresponding HSS and further incubated for 30 min. The localization of proteins bound to the plasmids was examined by ChIP analysis as described in Figure 2, except for using primer sets at X and Z, proximal and distal to the origin, respectively. Antibodies used for ChIP are indicated at the top of each panel. (D) Stable binding of Cdc45, Mcm2-7, and GINS to the plasmid both in the presence and absence of RecQ4. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted extracts as described in Figure 6A. After incubation in the mock- or RecQ4-depleted NPE, plasmid-beads were isolated and washed with the buffer containing various concentrations of NaCl. Proteins bound to the plasmids (plasmid) and in the extracts (extract) were analyzed by immunoblotting. (E) RecQ4 promotes unwinding of DNA in NPE. The bead-coupled plasmids (7.6 kbp) were incubated at 23°C for 30 min in RecQ4-depleted HSS, and then 2 vol of RecQ4-depleted NPE was added with and without a 720 nM N-terminal fragment of RecQ4 (N2) in the presence of 67 ng/μl aphidicolin. After 30 min incubation, the plasmids were isolated and treated with P1 nuclease at 38°C for 10 min. Total DNA isolated from the mixture was further digested with BamHI, and the purified DNA was separated by agarose gel electrophoresis. As a positive control, mock-depleted extracts were used for the unwinding assay (mock).](/cms/asset/6a708186-563d-4a3b-a989-f34d2aead138/kccy_a_1007003_f0006_oc.gif)
