Figures & data
Figure 1. LIF in delays contact inhibition. The size of HCECs in diameter was compared between cultures in SHEM and MESCM for 6 weeks (A) *P < 0.05, when compared to the size a week before in the same medium; #P < 0.05, when compared to that in SHEM at the same time point, n = 3). BrdU labeling of HCECs in different culture media at Day 7, 21, 35 and 42 was compared (B) *P < 0.05, **P < 0.01 when BrdU labeling in SHEM at the same time point was used as control; #P < 0.05, when BrdU labeling in ESCM at the same time point was used as control, n = 3). At Day 21, p120 was located at intercellular junctions in the cells cultured in SHEM (C). At Day 42, BrdU labeling was compared in all cultures (C). After 6 weeks of culture, the cell morphology, the junctional staining of p120, N-cadherin, β-catenin, ZO-1, F-actin, and Na-K-ATPase, and negative staining patterns of LEF1 and S100A4 were compared (D). Scale bar: 25 μm.
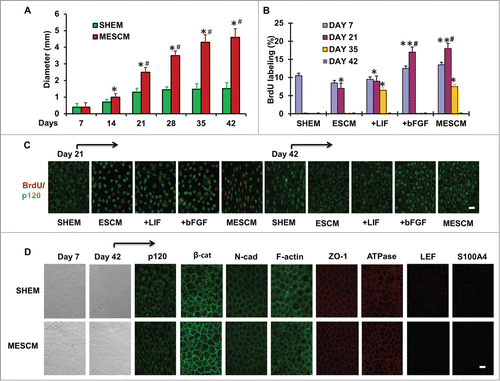
Figure 2. LIF promotes over-expression of ESC and NC markers. Transcript expression of ESC markers (A) and neural crest cell markers (B) was measured by qRT-PCR after 21 days of culturing of HCECs in different media (*P < 0.05, **P < 0.01 and ***P < 0.001, compare with that of SHEM group as the control, n = 3). The immunostaining was performed at Day 21 for expression of Sox2, Oct4, Nanog, KLF4, Nestin, p75NTR and FOXD3 (C). Scale bar: 25 μm.
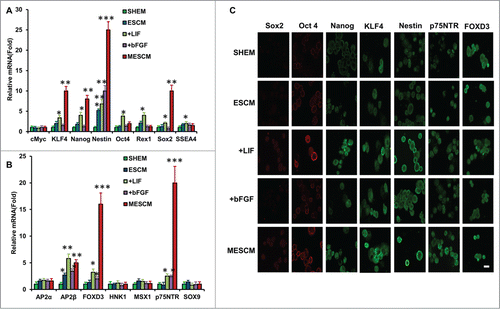
Figure 3. LIF or bFGF or combination does not activate BMP signaling. Expression of BMP ligands and receptors (A), and BMP downstream genes, ID1-4 (C), was measured by qRT-PCR (*P < 0.05 when compared to the corresponding SHEM group, n = 3) in HCECs at Day 21 cultured in different media. Immunofluorescence staining of pSMAD1/5/8 and pNF-κB was performed to examine the canonical BMP signaling and non-canonical BMP-NFκB signaling in HCECs (B). Scale bars: 25 μm.
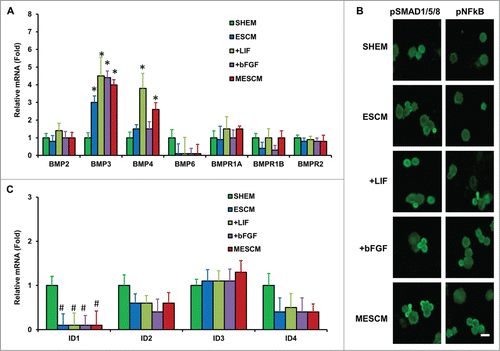
Figure 4. LIF-JAK1-STAT3 signaling in HCECs. The expression of mRNA of JAKs and STAT3 was measured at Day 21 in different cultures by qRT-PCR (A) *P < 0.05 when the corresponding SHEM group as the control; #P < 0.05 when the corresponding ESCM group as the control, n = 3). The immunostaining of Tyr705-phosphorylated form of STAT3 in HCECs cultured in ESCM+LIF and MESCM for 3 h was obtained from the confocal microscope (B). Scale bars: 25 μm.
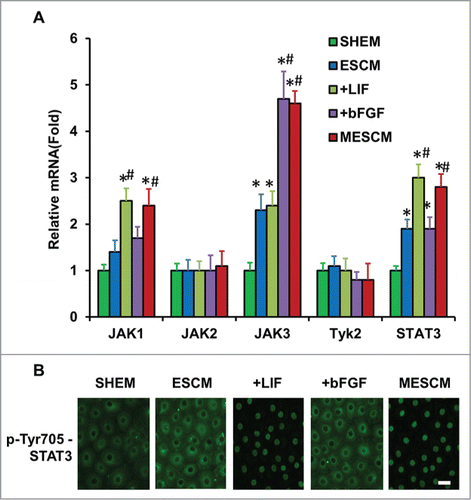
Figure 5. LIF delays contact inhibition and overexpression of ESC and NC markers. At Day 21, scRNA, JAK1 or STAT3 siRNA was added to the cultures for 48h and BrdU labeling was measured by immunostaining (A). At Day 35, JAK1 or STAT3 siRNA was added to the cultures for 48 h and BrdU labeling was measured by immunostaining (B) *P < 0.05 when compared to the scRNA group). The expression of ESC and NC markers was measured 48 h after JAK1 and STAT3 siRNA knockdown in HCECs cultured in ESCM+LIF (C) *P < 0.05 using ESCM group as the control, #P < 0.05 using ESCM+LIF group as the control) and ESCM+bFGF (D) *P < 0.05 using ESCM group as the control, #P < 0.05 using ESCM+bFGF group as the control) at Day 21 by qRT-PCR. Scale bars: 25 μm.
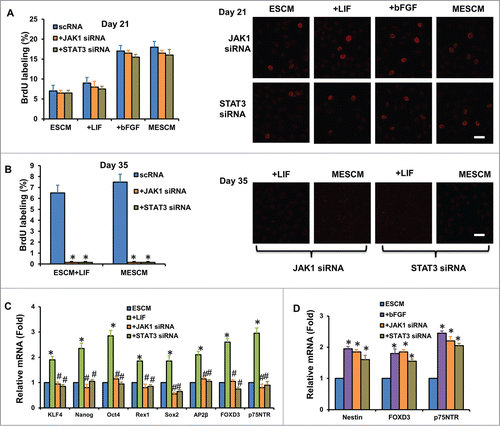
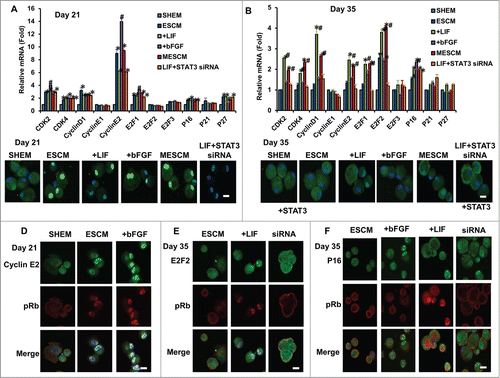
Figure 6. Association of expression of cell cycle related genes and contact inhibition. The mRNA expression of cell cycle related genes at Day 21 (A) and Day 35 (B) were measured by qRT-PCR (*P < 0.05 when the corresponding SHEM group as the control; #P < 0.05 when the corresponding ESCM group as the control, n = 3). The immunostaining of p-Rb (Ser807/811) at Day 21 and Day 35 in HCECs cultured in different media (C). The double immunostaining of p-Rb (Ser807/811) and Cyclin E2 (D), E2F2 (E) and p16INK4a (F) at Day 21 or Day 35 was obtained from the confocal microscope. Scale bars: 25 μm.