Figures & data
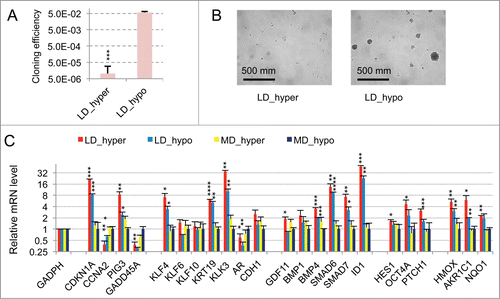
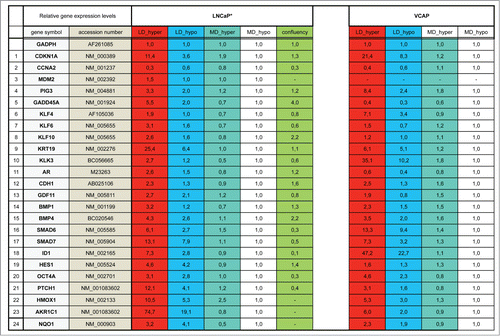
Figure 1. Characterization of a specific genetic expression signature of dormant LNCaP* cells. (A) RT-qPCR analysis of the variations of mRNA species levels according to cell culture conditions. Relative mRNA levels were averaged using the Averaging Ratio Method (ARM), with all mRNA levels reported to those measured under MD_hypo conditions in parallel experiments. Three to 5 independent experiments were performed with cells cultured in parallel under LD_hyper, LD_hypo, MD_hypo and 2 under MD_hyper conditions. (B) Cell cycle analysis of LNCaP* cells cultured under exponential growth (MD_hypo) or highly confluent conditions (confluency; see Experimental procedures). Cells were analyzed by Flow Cytometry after propidium iodide staining. A two-step gating procedure using Side SCattering-Height versus Forward SCattering-Height (data not shown) followed by pulse Area vs. pulse Width FLuorescence (FL2-A-versus FL2-W) was performed to eliminate cell doublets. Gated region is indicated on the FL2-A-versus FL2-W dot plot representation. Next to dot plots are fluorescence histograms. One representative experiment is shown. (C) Comparison of the patterns of mRNA level variations between confluent and dormant cells. Relative mRNA levels were averaged using the Normalized Averaging Method (NAM with levels under MD_hypo conditions taken as unity) from 2, 3, and 5 or 6 cell cultures experiments for confluent, dormant (LD_hyper conditions) and exponentially growing (MD_hypo) cells, respectively. Data are presented as mean ± s.d., with statistical significance indicated for the differences in mRNA levels between cells cultured under exponentially growing (MD_hypo) and under dormancy-inducing (LD_hyper) or permissive (LD_hyper) conditions (A), or between cells cultured under dormancy-inducing and confluent conditions (B); *, P < 0.1; **, P < 0.05; ***, P < 0.01; ****, P < 0.002. See the Materials and Methods section for more details on the averaging methods.
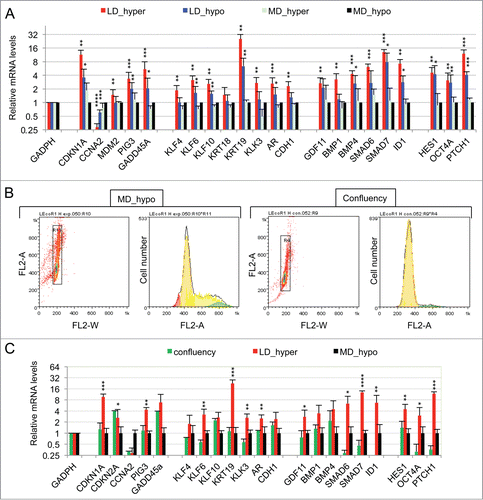
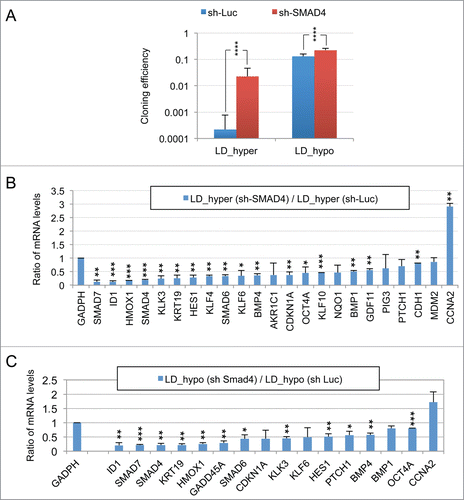
Figure 2. Implication of SMAD signaling in LNCaP* dormancy. (A) Western Blot analysis of total smad7 expression (endogenous human SMAD7 and transduced murine Smad7) in cell populations transduced with pTRIP-control and pTRIP-mSmad7 cultured under exponentially growing (MD_hypo) or dormancy inducing (LD_hyper) conditions. Cells were grown in the presence of doxycycline to induce pTRIP expression and a polyspecific monoclonal antibody was used. The arrow indicates the common expected size for SMAD7 and Smad7 proteins (46 kDa) while a star indicates bands resulting from non-specific antibody hybridizations. Two identically processed parts of the same gel have been regrouped for the figure. (B) Regulation of mRNA species accumulation under dormancy-inducing conditions by ectopic Smad7 overexpression. Ratios of mRNA species levels measured by RT-qPCR between Smad7 and control-transduced cell populations cultured under LD_hyper conditions were derived from 2 couples of cell populations. Statistical significance of the effects of Smad7 overexpression is indicated. (C) Regulation of mRNA species accumulation by BMP4 and Activin α signaling in cells cultured under MD_hypo conditions. Relative mRNA levels determined by RT-qPCR were averaged with NAM (with levels under MD_hypo conditions taken as unity) from 2 and 4 independent pTRIP-BMP4 and pTRIP-control transduced cell populations respectively, cultured under MD_hypo conditions in the presence of doxycycline and activin α at 50 ng/ml as indicated. Statistical significance of the difference in mRNA levels between cells stimulated by BMP4 plus activin α and control cells is indicated. D) Regulation of mRNA species accumulation by activin α signaling in LNCaP* cells cultured under MD_hypo conditions. Relative mRNA levels were measured by RT-qPCR from 2 control and 2 treated cell populations respectively. Note that mRNA level of ID1, a target gene of BMP signaling, was decreased by activin α addition. Data are presented as mean ± s.d., with significance of mRNA levels variations indicated as in .
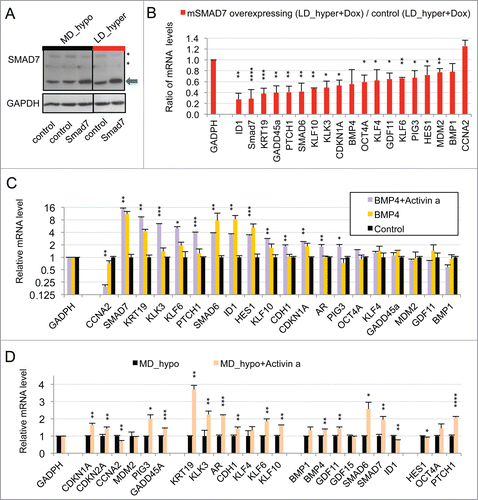
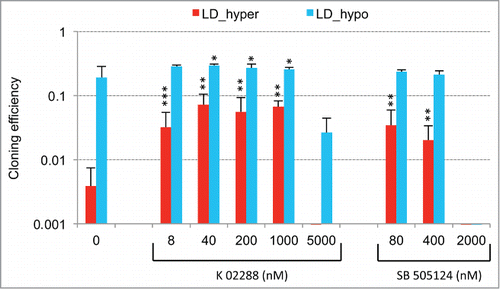
Figure 3. Modulation of dormancy by glutathione (GSH). (A) Glutathione supplementation reverse the effect of hypertonicity on the cloning efficiency of LNCaP* cells. Statistical significance of the effects of glutathione addition is indicated. (B) Pictures of representative cloning efficiency assays for the indicated cell culture conditions. (C) LNCaP* cell cloning inhibition by Buthionine Sulfoximine (BSO) under hypotonic conditions and its reversal by glutathione supplementation. Cloning efficiency of LNCaP* cells cultured in hypotonic conditions (LD_hypo) in the presence of BSO and/or Glutathione as indicated were determined. Data are averaged from 2 independent cell experiments and are presented as mean ± s.d., with statistical significance indicated for the effects of glutathione supplementation. (D) Pictures of cells after 13 d of culture under LD_hypo conditions in the presence of none, GSH 8mM, BSO 0.25 mM and BSO plus GSH. (E) Glutathione supplementation reverses the gene expression signature of dormancy. Relative mRNA levels were calculated as in as the ratio of mRNA levels species measured in LNCaP* cell populations cultured under LD_hyper conditions in the presence of 4 mM glutathione and in MD_hypo conditions. Data are averaged from 3 cell culture experiments. Relative mRNA levels under LD_hyper and LD_hypo conditions are those of . Statistical significance of the differences between dormant and glutathione-treated cells is indicated as in .
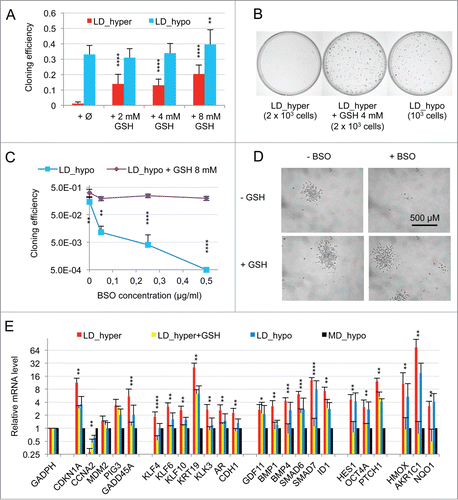
Figure 4. Oxidative stress mediated by hydrogen peroxide cooperates with BMP4/activinα signaling to induce the dormancy gene expression signature in LNCaP* cells cultured at middle cell density in hypotonic medium (MD_hypo). (A) Effect of H2O2-induced oxidative stress on mRNA species accumulation in LNCaP* cells cultured under MD_hypo conditions. Inset: Dose response of the cloning efficiency of LNCaP* cells to H2O2. Data are average of 2 cloning efficiency assays made with 103 and 2 × 103 cells in 6 cm diameter dish, each in duplicate. Histogram: Effects of hydrogen peroxide treatment on mRNA species accumulation. Relative mRNA species levels were averaged with NAM from 2 parallel cell culture experiments. Statistical significance of the effects of H2O2 addition is indicated. All Data are presented as mean ± s.d., with significance indicated as in . (B) Comparison of the mRNA accumulation patterns generated by activation of BMP4/ activin α signaling plus H2O2 treatment and by dormancy. Relative mRNA levels were averaged with NAM from 2 independent cell populations transduced with pTRIP-BMP4 and cultured under MD_hypo conditions in the presence of doxycycline (Dox; 0.5 μg/ml) + activin α (50 ng/ml) + H2O2 100 μM, 4 independent cell populations transduced with pTRIP-control and cultured under LD_hyper conditions in the presence of doxycycline and 4 to 6 independent cell populations transduced with pTRIP-control and cultured under MD_hypo conditions in the presence of doxycycline. Data were calculated using the normalized averaging method and are presented as mean ± s.d., and stars indicate statistical significance of mRNA level variations compared to basal levels measured for pTRIP-control transduced cell populations cultured under MD_hypo conditions in the presence of doxycycline.
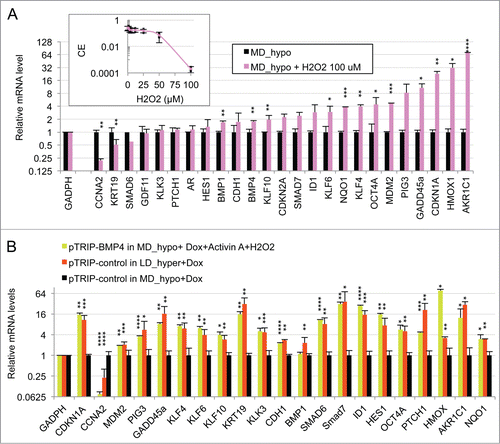
Figure 5. Activation of SMAD signaling at low cell density and contribution of ROS signaling. (A) Smad7 overexpression changes the gene expression profile of LNCaP* cells under LD_hypo conditions. Ratios of mRNA species levels measured by RT-qPCR in pTRIP-mSMAD7 and pTRIP-control transduced cell populations cultured under LD_hypo conditions were averaged from 2 experiments using independent cell populations. Stars indicate significance of the decreased mRNA species accumulation upon murine Smad7 overexpression. (B) Glutathione supplementation reduces the activation of SMAD signaling at low density. Relative mRNA levels measured by RT-qPCR under LD_hypo, LD_hypo+glutathione and MD_hypo conditions were averaged from 4–5, 2 and 7–9 cell culture experiments, respectively. Stars indicate statistical significance of the difference in mRNA levels between cells cultured in LD_hypo and LD_hypo+glutathione. (C) Lack of additive effects between Smad7 overexpression and glutathione treatment on cloning efficiency. Cloning efficiency of pTRIP-control and pTRIP-mSmad7 transduced cell populations were measured under the indicated cell culture conditions. Data were averaged from 2 to 5 experiments made with 2 independent control and 2 independent Smad7 transduced cell populations processed in parallel. Stars indicate significance of the differences in cloning efficiency. Data are presented as mean ± s.d.
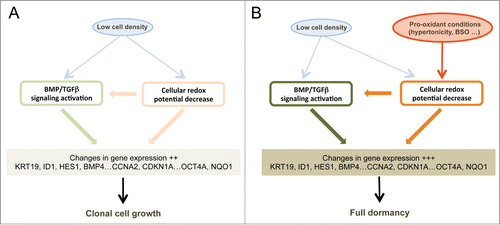
Figure 6. Schematic representation of the signaling pathways involved in cell dormancy. (A) Cell signaling pathways activated by low cell density under clonogenic conditions (i. e. in isotonic or slightly hypotonic culture medium). Under these conditions, a mild activation of BMP/TGFβ (, and Fig. S4C) and redox imbalance ( and ) signaling pathways is observed, redox imbalance also contributing to the activation of BMP/TGFβ signaling (). (B) Reinforcements of these signaling pathways under dormancy-inducing conditions in slightly hypertonic medium. The main effect of hypertonicity is to amplify redox imbalance and BMP/TGFβ signaling (, and Fig. S4B and C) thus increasing the expression level of the dormancy gene expression signature that is already activated in cells plated at low density but in hypotonic or isotonic medium.