Figures & data
Figure 1. KDM4D region spanning 350–474 amino acids binds PAR in vitro. PAR-binding assay with 6xHis tagged KDM4D full-length (FL) protein (523aa) (A), deletion mutants: N-terminal (1–350aa) (B), GST-tagged C-terminal (350–523aa) (C), truncated C-terminal (1–474aa) and internal deletion 350–474aa (D), and 6xHis tagged KDM4D-4M mutant (contains 4 mutations: E357A, R450A, R451A and R455A) (E). 6xHis-Rpn8, GST-only and BSA are used as negative controls and H3 as a positive control. Right: schematic representation of KDM4D mutants. IB: Immunoblot. 32P: radiolabelled PAR.
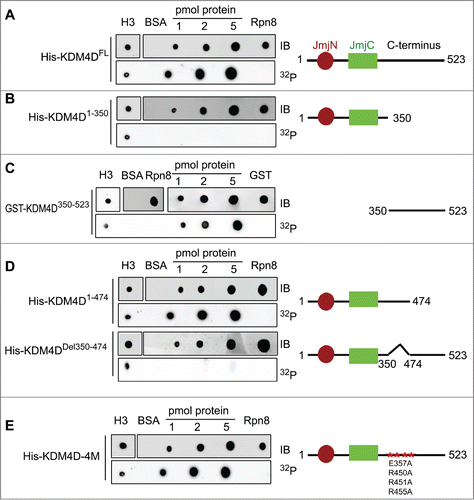
Figure 2. KDM4D PAR-binding region is essential for its recruitment to laser-microirradiated sites. Representative cells showing the localization of EGFP-KDM4D-WT EGFP-KDM4D1-474aa and EGFP-KDM4DΔ350-474aa fusions before and 5 minutes after the induction of laser-microirradiation to a single region, marked with a white arrow. Each cell is representative of at least 20 different cells. The graph shows the increase in the relative fluorescence intensity of KDM4D fusions at laser-microirradiated sites. Error bars represent SD of 10 different cells.
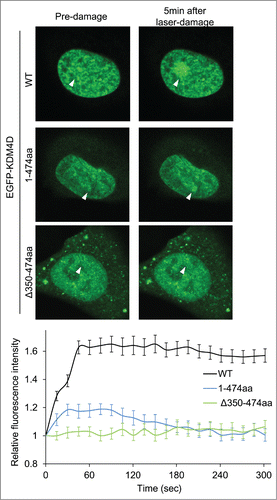
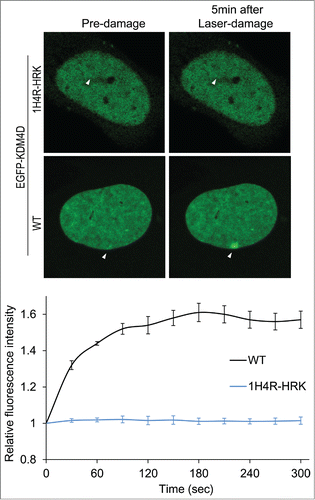
Figure 3. KDM4D-RNA interactions are essential for KDM4D accumulation at DNA damage sites. Representative cells showing the localization of EGFP-KDM4D-WT and EGFP-KDM4D-1H4R-HRK fusions before and 5 minutes after the induction of laser-microirradiation to a single region, marked by a white arrow. Each cell is representative of at least 20 cells. The graph shows the increase in the relative fluorescence intensity of EGFP-KDM4D-WT and EGFP-KDM4D-1H4K-HRK at laser-microirradiated sites. Error bars represent SD of 10 different cells.